Figure 5.
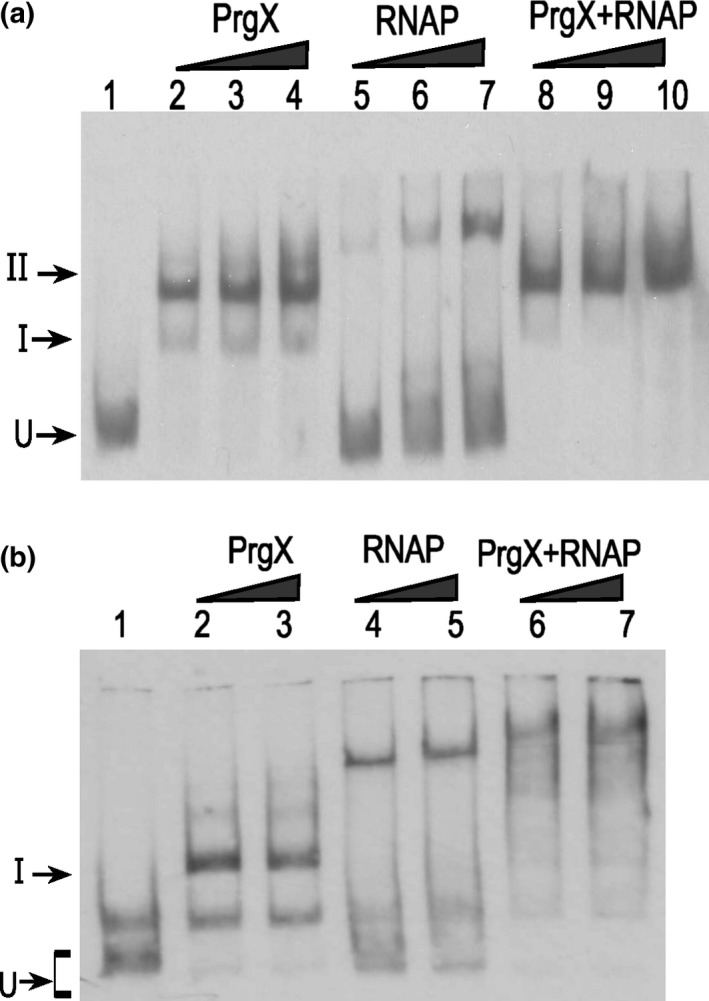
PrgX and E. faecalis RNA polymerase (RNAP) compete for binding at prgQ promoter. Electrophoretic mobility shift assays were performed using 8 fmol of digoxigenin‐labeled DNA probes and various amounts of purified PrgX and RNAP. PrgX binds to DNA probe containing both operator XBS1 and XBS2 (LT template) and forms two‐shifted complexes (band I and II). “U” indicates the position of unbound probe. The components in each lane are as indicated in the figure. (a). PrgX hinders RNAP binding to prgQ promoter. LT DNA was first incubated with RNAP for 10 min at RT before addition of PrgX to the reactions. Untagged PrgX was cleaved from GST‐PrgX. PrgX concentration used: lanes 2 and 8: 19 nmol L−1; lanes 3 and 9: 38 nmol L−1; lanes 4 and 10: 72 nmol L−1. RNAP concentration used: lanes 5 and 8: 60 nmol L−1; lanes 6 and 9: 70 nmol L−1; lanes 7 and 10: 150 nmol L−1. (b). PrgX and RNAP form a stable complex on LT DNA which has mutations in the XBS2 site. In these reactions, PrgX and RNAP were incubated with probe DNA probes at RT for 15 min. PrgX concentration used: lanes 2 and 6: 18 nmol L−1; lanes 3 and 7: 54 nmol L−1. RNAP concentration used: lanes 4 and 6: 200 nmol L−1; lanes 5 and 7: 240 nmol L−1
