Figure 4.
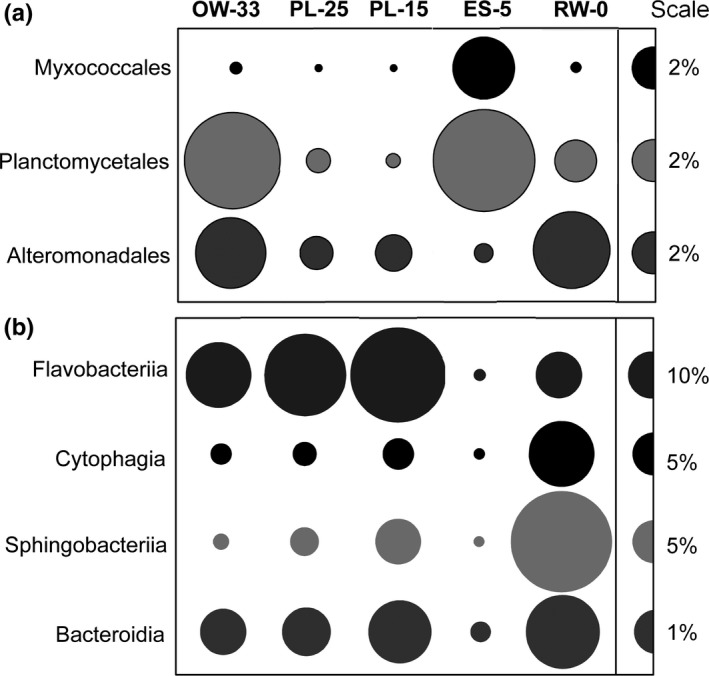
Relative abundance of bacterial taxa based on predicted gene annotations in assembled metagenomes: (a) bacterial orders predicted to be involved in phytoplankton turnover; and (b) four classes of the phylum Bacteroidetes. Abundance values are expressed as percentages of the corresponding genes out of the total annotated CDS in a given metagenome (with annotations based on ≥30% identity over ≥70% of the alignment length). The abundance values are proportionate to the bubble width. The scale for each row is indicated with a half‐bubble to the right, and its diameter corresponds to a percentage value shown with a number. The metagenome names provide information on sampling locations and water salinity (PSU; OW‐33, ocean water; PL‐25 and PL‐15, plume; ES‐5, estuarine; RW‐0, freshwater)
