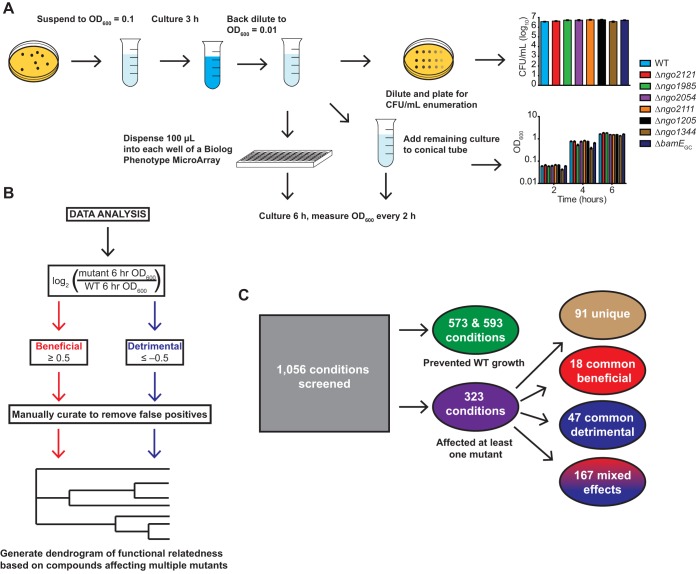
FIG 3.
Phenotype screening procedure. (A) Flowchart outlining the optimized phenotype screening procedure. Nonpiliated colonies of FA1090 (WT) and mutant bacteria were collected from GCB plates, suspended in GW medium to an OD600 of 0.1, cultured for 3 h in a 5% CO2 environment at 37°C, and back diluted to an OD600 of 0.01. Cultures were split for CFU enumeration, overall growth monitoring, and phenotype screening. Inoculum for screening plates was serially diluted 10-fold from −1 to −4, and 5 μl was spotted on GCB plates for enumeration. OD600 values of screening plates and cultures without compounds were measured every 2 h for each strain. Bars represent means ± SEM of cultures without compounds. WT, n = 17; Δngo2121 and Δngo1985 mutants, n = 6; Δngo2054, Δngo2111, Δngo1205, Δngo1344, and ΔbamEGC mutants, n = 11. (B) Flowchart outlining data analysis. Endpoint OD600 values were used to calculate a log2-transformed ratio of mutant to WT growth. Beneficial conditions were designated those that resulted in a ratio of ≥0.5. Detrimental conditions were those with a ratio of −0.5 or lower. Data were manually curated to remove false positives. Hits were used to construct a functional relatedness dendrogram of vaccine candidates. (C) Out of 1,056 conditions screened, 573 and 593 conditions prevented growth of WT in the first and second screenings, respectively. Three hundred twenty-three conditions had some effect on at least one mutant. Ninety-one conditions uniquely affected one mutant. Of the conditions with effects on multiple mutants, 18 conditions were commonly beneficial, 47 were commonly detrimental, and 167 had mixed effects. GCB, gonococcal base medium; GW, Graver-Wade liquid medium; OD600, optical density at 600 nm.