ABSTRACT
Agrobacterium tumefaciens is a rod-shaped bacterium that grows by polar insertion of new peptidoglycan during cell elongation. As the cell cycle progresses, peptidoglycan synthesis at the pole ceases prior to insertion of new peptidoglycan at midcell to enable cell division. The A. tumefaciens homolog of the Caulobacter crescentus polar organelle development protein PopZ has been identified as a growth pole marker and a candidate polar growth-promoting factor. Here, we characterize the function of PopZ in cell growth and division of A. tumefaciens. Consistent with previous observations, we observe that PopZ localizes specifically to the growth pole in wild-type cells. Despite the striking localization pattern of PopZ, we find the absence of the protein does not impair polar elongation or cause major changes in the peptidoglycan composition. Instead, we observe an atypical cell length distribution, including minicells, elongated cells, and cells with ectopic poles. Most minicells lack DNA, suggesting a defect in chromosome segregation. Furthermore, the canonical cell division proteins FtsZ and FtsA are misplaced, leading to asymmetric sites of cell constriction. Together, these data suggest that PopZ plays an important role in the regulation of chromosome segregation and cell division.
IMPORTANCE A. tumefaciens is a bacterial plant pathogen and a natural genetic engineer. However, very little is known about the spatial and temporal regulation of cell wall biogenesis that leads to polar growth in this bacterium. Understanding the molecular basis of A. tumefaciens growth may allow for the development of innovations to prevent disease or to promote growth during biotechnology applications. Finally, since many closely related plant and animal pathogens exhibit polar growth, discoveries in A. tumefaciens may be broadly applicable for devising antimicrobial strategies.
KEYWORDS: Agrobacterium, PopZ, cell polarity, chromosome segregation, cell division, growth polarity
INTRODUCTION
Agrobacterium tumefaciens, the causative agent of crown gall disease in flowering plants, has been studied extensively with regard to pathogenesis and its ability to transfer engineered DNA to plant cells using the type IV secretion system encoded by multiple vir genes. More recent observations of A. tumefaciens growth have revealed that this bacterium exhibits polar growth during elongation (1, 2). In A. tumefaciens, as well as in other Rhizobiales, peptidoglycan is inserted at the new poles created by cell division until the cell doubles in length. The peptidoglycan synthesis is then directed to midcell to enable septum formation and cell division. The growth patterning of A. tumefaciens suggests that both spatial and temporal regulation are necessary to restrict cell wall biosynthesis to the pole during elongation and to midcell during cell division.
Many of the genes encoding canonical proteins known to function in cellular elongation, including MreB, MreC, MreD, RodA, PBP2, and RodZ, are absent from the A. tumefaciens genome; however, the genes encoding the cell division machinery are well conserved (1, 3, 4). Remarkably, studies on the dynamics of FtsZ and FtsA suggest that both proteins have an expanded role contributing to the regulation of peptidoglycan biosynthesis not only at the midcell but also at the growth pole (1, 5, 6). FtsZ and FtsA persist at the growth pole, and the delocalization of these proteins from the growth pole coincides with the transition of the growth pole to an inert, old pole (6). Once delocalized from the pole, FtsZ and FtsA sequentially appear at midcell prior to the initiation of peptidoglycan biosynthesis at midcell (5). When the bacterium divides, FtsZ and FtsA are retained at the new cell poles formed from the division plane. These observations have led to the suggestion that a combination of cell division machinery and novel proteins is required for polar elongation (3).
What types of novel proteins may function in polar elongation? In many diverse bacteria, poles can act as a subcellular hub for proteins involved in cell development (7). Among the alphaproteobacteria, polar organizing proteins are best described in Caulobacter crescentus. C. crescentus PopZ (PopZCc) localizes to the new pole, where it interacts with ParB and other chromosome partitioning machineries to segregate the chromosome (8–10). PopZCc also localizes to the old pole, where it does not tether the chromosome but rather functions in the localization of polar regulatory proteins, including histidine kinases which function in cell cycle control (8, 10). A. tumefaciens PopZ (PopZAt) does not exhibit bipolar localization but rather is found strictly at the new pole (5) and accumulates in ectopic poles formed in a mutant with cell division defects (11). Based largely on its strict localization to growing poles, the polar organizing protein, PopZ, has been identified as a candidate protein for promoting polar growth in A. tumefaciens (5, 11).
Here, we characterize the role of PopZ in the regulation of growth patterning, polarity, and cell division of A. tumefaciens. Surprisingly, we find that polar elongation and biosynthesis of polar structures continues in the absence of PopZ; however, the loss of PopZ causes morphological defects indicative of a block in cell division. Furthermore, the cell division proteins FtsZ and FtsA are often misplaced, leading to the production of asymmetric sites of constriction and a broad cell length distribution. A high proportion of cells lacking PopZ are devoid of DNA, suggesting a conserved function in chromosome segregation. Together, these results indicate that PopZ has a critical role in the regulation of cell division despite its strict growth pole localization pattern.
RESULTS
Loss of PopZ causes cell division defects, ectopic pole formation, and minicell formation.
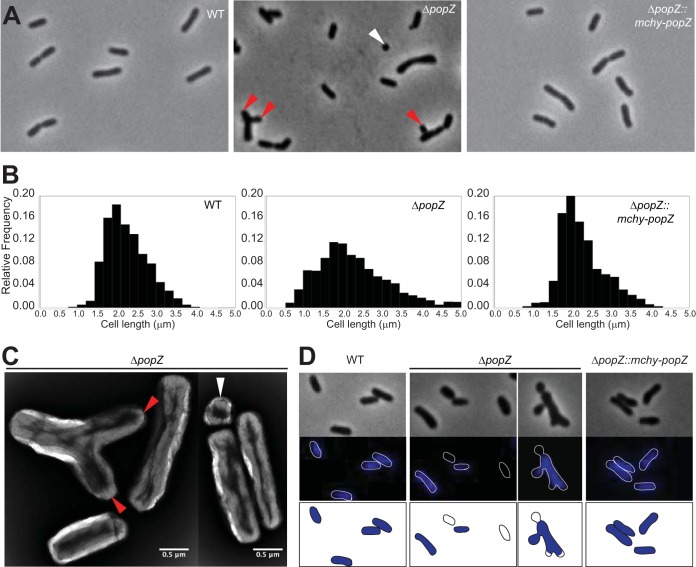
To determine the role of PopZ in A. tumefaciens, a ΔpopZ deletion strain was used in which the native gene of popZ was replaced with a genetic cassette bearing spectinomycin resistance (12). The ΔpopZ cells have a doubling time that is approximately 40% longer than that of wild-type cells (∼167 min for ΔpopZ cells compared to ∼120 min for wild-type cells [12]) and display a range of morphological defects, including ectopic poles, bulged side walls, and abnormal cell lengths (Fig. 1). In wild-type C58 C1 (WT) cells, less than 1% of the population displays branches or bulges, while these phenotypes are observed in 40% of the ΔpopZ population.
FIG 1.
Analysis of morphology, cell length, and DNA content of the popZ deletion strain. (A) Comparison of phase-contrast images of wild-type, ΔpopZ, and ΔpopZ::popZ-mchy strains grown to exponential phase in ATGN media. The ΔpopZ culture contains a high proportion of small cells (<1.5 μm in length; white arrowhead) and branched cells with ectopic poles (red arrowheads). (B) Cell length distributions of WT (left; n = 926), ΔpopZ (center; n = 1,664), and ΔpopZ::popZ-mchy (n = 839) cells are shown. (C) Transmission electron micrographs of nano-tungsten-stained ΔpopZ cells. The deletion of popZ results in an increased cell length distribution, including very small cells (white arrowhead) and cells with ectopic poles (red arrowheads). (D) DAPI staining reveals the presence of anucleate cells in the ΔpopZ population. Phase (top) and fluorescent (middle) images of representative DAPI-stained wild-type, ΔpopZ, and ΔpopZ::popZ-mchy cells are shown. Outlines are provided to indicate cell location in fluorescent images. Schematics of DAPI-stained cells are provided in the bottom panel.
The morphological defects observed in the ΔpopZ mutant result in a broader cell length distribution, including increases in both short and long cells (Fig. 1B). In the WT, 94% of the cells are between 1.5 and 3.5 μm in length, while only 70% of ΔpopZ cells fall into this range. Remarkably, we observed a marked increase in the percentage of cells with lengths less than 1.5 μm in ΔpopZ cells (29% ΔpopZ cells compared to 6% in WT cells). To determine if the small cells contain DNA, we stained ethanol-fixed cells with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) to visualize the DNA (Fig. 1D). Many of the small cells lack DNA and appear to arise from cell divisions near the pole prior to the completion of chromosome segregation. Furthermore, we observed cells of typical cell lengths (between 1.5 and 3.5 μm) that lack DNA. These observations are consistent with a role for PopZ in chromosome segregation, as described for Caulobacter crescentus (8, 9). Notably, replacement of the native popZ gene with a translational mcherry-popZ fusion mimics wild-type cell morphologies, length distributions, and DNA content, indicating that mChy-PopZ is functional (Fig. 1).
PopZR2-R3-sfGFP localizes to the new pole but does not restore morphology defects.
Previous reports show that in A. tumefaciens, PopZ localizes to the growing pole during cell elongation and arrives at the two newly formed poles just after cell division (5). We observed the same localization pattern using a low-copy-number plasmid with an isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible popZ-sfgfp in WT (Fig. 2A, top) and ΔpopZ (Fig. 2A, bottom) backgrounds. This construct was able to rescue the morphological defects of the popZ deletion, indicating that PopZ-superfolder green fluorescent protein (sfGFP) is functional (Fig. 2B).
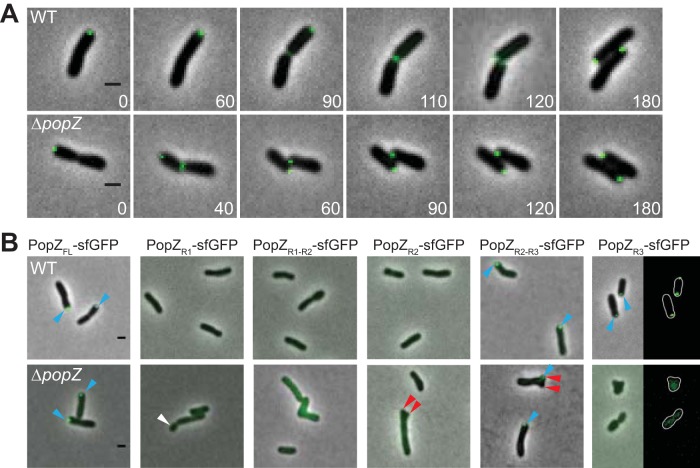
FIG 2.
Polar localization of PopZ requires domains R2 and R3. (A, top) Time-lapse microscopy of WT cells expressing full-length PopZ-sfGFP (PopZFL-sfGFP) reveals a new pole-to-midcell localization pattern. (Bottom) PopZFL-sfGFP exhibits a similar localization pattern in the ΔpopZ strain. (B) Representative images of the localization patterns of full-length and truncated versions of PopZ-sfGFP in wild-type and ΔpopZ cells are shown. In wild-type cells, PopZ truncations containing domain R3 (PopZFL-sfGFP, PopZR2-R3-sfGFP, and PopZR3-sfGFP) localize to growth poles (blue arrowheads). In the absence of PopZ, only PopZFL-sfGFP and PopZR2-R3-sfGFP localize to growth poles (blue arrowheads). Truncated forms of PopZ do not complement the morphological defects of ΔpopZ cells, as indicated by the presence of small cells (white arrowheads) and ectopic poles (red arrowheads). All scale bars are 1 μm.
The PopZ protein is conserved among many alphaproteobacteria and typically contains three domains: R1 (amino acids [aa] 1 to 25) is an N-terminal domain containing an alpha helix, R2 (aa 26 to 258) is the central proline-rich domain, and R3 (aa 259 to 333) is the C-terminal domain containing multiple alpha helices (13, 14). Here, we sought to determine which of the PopZ domains are necessary for proper subcellular localization. Thus, we constructed truncations of PopZ fused to sfGFP at their C termini, which are present in both WT and ΔpopZ backgrounds (see Fig. S1 in the supplemental material). In a WT background, we noted that the R3 domain was necessary and sufficient for polar localization (Fig. 2B, top). In the absence of native PopZ, however, the R3 domain was not sufficient for polar localization, and instead proper subcellular localization required both the R2 and R3 domains (Fig. 2B, bottom). Although the PopZ R2-R3 domain is sufficient for polar localization of sfGFP, it does not rescue the morphological defects, suggesting a functional role for the R1 domain in A. tumefaciens. In C. crescentus, the conserved R1 domain of PopZ is required for interactions with chromosome portioning proteins ParA and ParB, and these interactions are required for proper cell division and chromosome segregation (15). Indeed, none of the PopZ truncations could rescue the morphological defects in the ΔpopZ strain (Fig. 2B), indicating that the three domains each have important contributions to PopZ function in A. tumefaciens.
PopZ is not required for unipolar polysaccharide biosynthesis or adhesion to abiotic surfaces.
The subcellular localization pattern of PopZ in A. tumefaciens indicates that PopZ functions specifically at the new pole. The new pole is the site of unipolar polysaccharide (UPP) biosynthesis, flagellum biosynthesis, and peptidoglycan biosynthesis during cellular elongation (16, 17). In order to determine if PopZ contributes to any of these processes, we examined the biosynthesis of each structure in WT and ΔpopZ cells.
First, we determined the role of PopZ in the production and function of UPP. The UPP was detected using wheat germ agglutinin conjugated to Alexa Fluor 488 (WGA-488) (Fig. 3A). In WT, ΔpopZ, and ΔpopZ::mchy-popZ cell populations, UPP was detected at the pole in approximately 20% of the cells. In ΔpopZ cells, the UPP is typically found at one pole; however, some cells with ectopic poles have UPP at multiple poles (Fig. 3A). In order to assess if the UPP retained its function in surface attachment, we completed short-term binding assays (Fig. 3B). Surprisingly, after 1 h we observed a significant increase in the number of cells bound to a glass coverslip in the absence of popZ. Remarkably, the ΔpopZ cells seem to form large aggregates that tightly bind to the glass coverslip. In addition, the loss of popZ results in a significant increase in biofilm formation after 48 h (Fig. 3C). While the biosynthesis and localization of the UPP do not seem to be disrupted in ΔpopZ cells, these cells do exhibit increased surface attachment. It is possible that the quantity of UPP has increased due to cell cycle perturbations, causing the cells to produce UPP for a longer duration, that adhesive and cohesive properties of the UPP have been modified, or that the production of another polysaccharide which contributes to surface attachment has been altered. Indeed, the A. tumefaciens genome indicates the presence of at least six biosynthetic pathways for polysaccharides, including cellulose and succinoglycan, which may contribute to attachment and biofilm formation (16), and the observation of large aggregates formed by the ΔpopZ cells is reminiscent of aggregates formed when cellulose is overexpressed (18).
FIG 3.
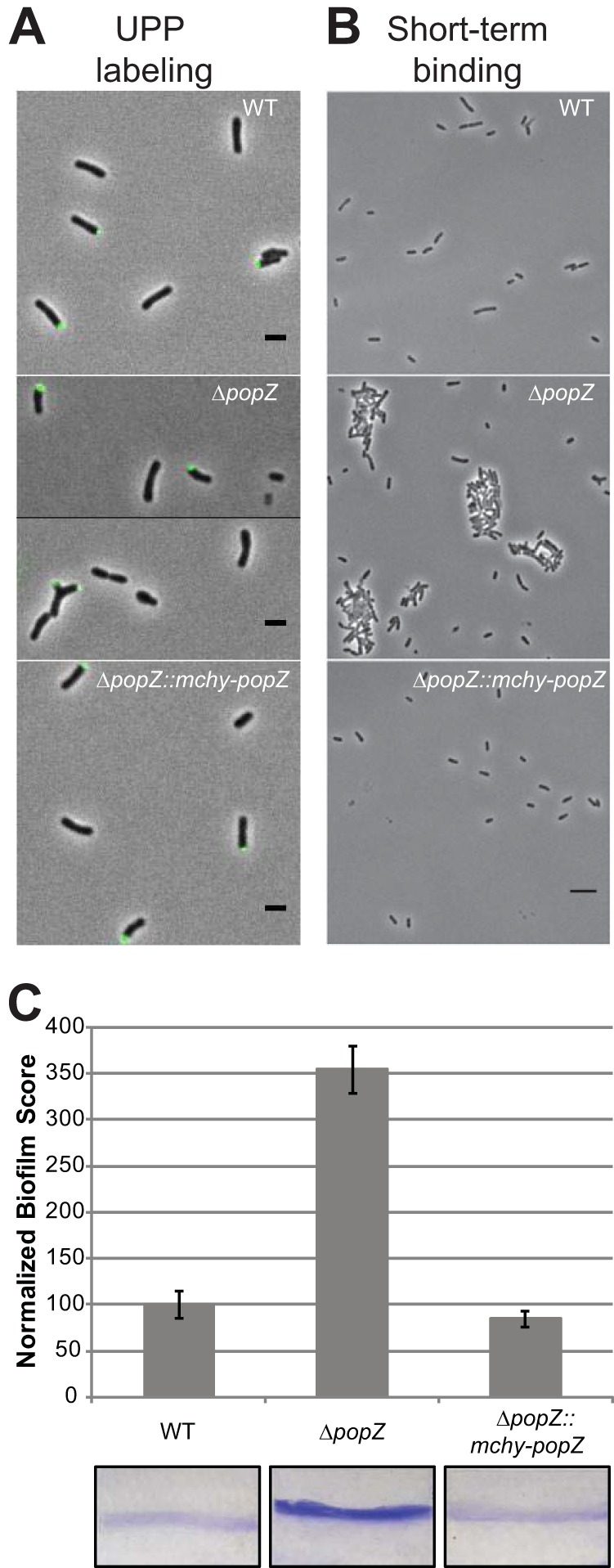
Analysis of biofilm formation in wild-type and ΔpopZ mutant strains. (A) UPP production and placement was identified by the binding of Alexa Fluor 488-labeled wheat germ agglutinin to cells on 1.5% agarose pads. In all three strains, approximately 20% of individual cells have a detectable UPP. Scale bars, 2 μm. (B) Short-term binding was evaluated after 1 h of attachment to glass coverslips. Scale bar, 5 μm. (C) Strains were assayed for biofilm formation on vertical plastic coverslips immersed in ATGN medium. Coverslips were removed after 48 h of incubation at room temperature and rinsed to remove any loosely associated cells. Adherent biomass was determined as the absorbance of solubilized crystal violet (A600), and the optical density of the planktonic culture (OD600) was measured. Biofilm scores were calculated as the A600/OD600 ratio, and data were normalized to WT values. Data shown are the means from two independent experiments completed in triplicate. Error bars are the standard errors of the means. Representative coverslips prior to crystal violet solubilization are shown for each strain.
Flagellum placement is independent of PopZ.
To determine the role of A. tumefaciens PopZ in the production of flagella, a yellow fluorescently tagged basal body protein (FliM-YFP) was used to track the position of flagella relative to mChy-PopZ (Fig. 4A). We observe that FliM-YFP foci are found near the pole containing mChy-PopZ, consistent with previous reports of subpolar flagella in A. tumefaciens (19). Time-lapse microscopy illustrates that as the cell elongates, the distance between the pole and FliM-YFP increases, indicating that the flagella remain fixed as new cell wall material is inserted at the growth pole. Indeed, the change in FliM-YFP distance from the pole is highly correlated with the increase in cell length (Fig. 4B). When FliM-YFP is expressed in ΔpopZ cells, FliM-YFP is observed in subpolar foci (Fig. 4C). The distance of the FliM-YFP foci from the pole increases as the cells elongate, suggesting the basal bodies of the flagella are properly assembled in the ΔpopZ cells, which is consistent with observations of motile cells in planktonic culture (data not shown) despite a slight defect in motility in swim plate assays (Fig. S2) due to defects in cell morphology and division. Furthermore, the observation that the distance of basal bodies from the poles increases as the cells get longer suggests that ΔpopZ cells are continuing to insert newly synthesized peptidoglycan at the growth pole, and this polar organizing protein may not be involved in organizing polar growth.
FIG 4.
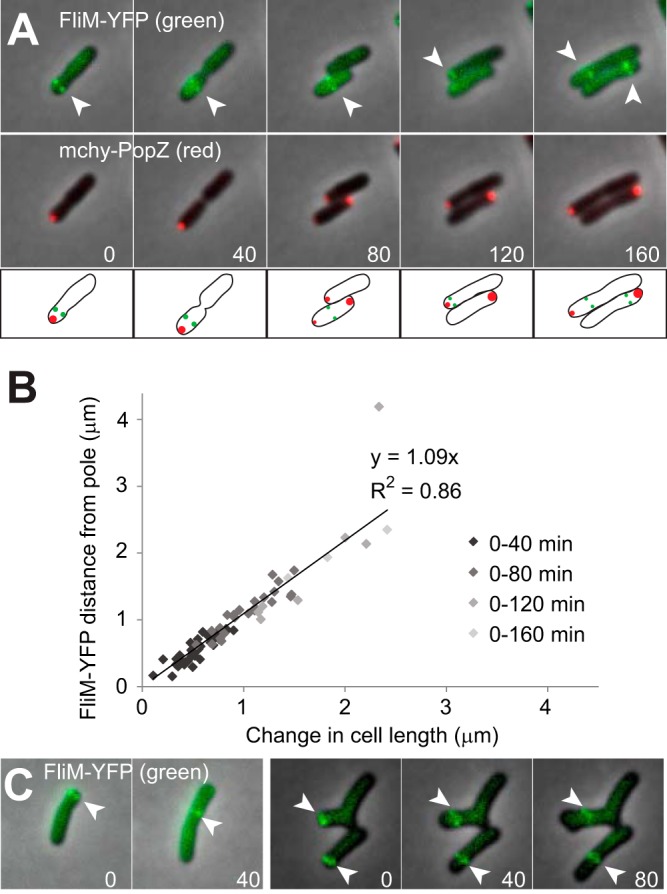
Analysis of flagellum localization in wild-type and ΔpopZ mutant strains. (A) Flagellar basal bodies were fluorescently labeled by expressing FliM-YFP, and their localization patterns (green) were observed by time-lapse fluorescence microscopy in cells that also express mChy-PopZ (red). FliM-YFP foci are indicated by arrowheads. Fluorescence images are overlaid on a phase-contrast background, and time (in minutes) is shown in the lower corners of images. Schematics are provided below the images. (B) For cells shown in panel A, a scatterplot compares the increase in cell length to the increase in the distance of the FliM-YFP foci from the cell poles. A total of 34 cells were measured in 2 independent experiments, and the slope of the linear regression and associated R-squared value is shown. There are fewer data points for later time points because most cells divided during the time course. (C) Fluorescence localization of FliM-YFP (green) in ΔpopZ cells.
Polar growth is retained in the absence of PopZ.
Due to the predicted molecular scaffolding properties of PopZ, as well as its localization to the growth pole, PopZ has been hypothesized to function in unipolar growth of A. tumefaciens (5, 11). While our tracking of flagellar basal bodies suggests that polar insertion of peptidoglycan continues in the ΔpopZ cells, we used fluorescent d-amino acids (FDAAs) to probe the growth pattern in the presence and absence of PopZ. In WT A. tumefaciens, FDAAs are preferentially incorporated into the muropeptide stem of peptidoglycan and reveal the subcellular regions exhibiting active peptidoglycan synthesis (2). The typical FDAA labeling pattern of A. tumefaciens is consistent with three growth stages: (i) polar peptidoglycan synthesis during cellular elongation, (ii) a transition stage in which polar elongation is terminated and midcell peptidoglycan synthesis is initiated, and (iii) strict midcell peptidoglycan synthesis enabling new growth-active poles to form following cell division (2). In the absence of PopZ, polar growth is retained across three morphology types: (i) rod shaped, (ii) small, and (iii) branched/bulged (Fig. 5). When ΔpopZ cells retain their rod shape and typical cell length (1.5 to 3.5 μm), FDAA labeling indicates that peptidoglycan synthesis occurs at the pole and midcell (Fig. 5A to D). Twenty-nine percent of ΔpopZ cells are shorter than 1.5 μm in length, and the vast majority of these cells do not label with FDAAs (Fig. 5G). Dual labeling with FDAAs and DAPI staining reveals that 95% (98/103) of small cells lacking active peptidoglycan biosynthesis also lack DNA (Fig. S3). Among the small cells with FDAA labeling, unipolar and bipolar localization was observed (Fig. 5E and F). Forty-one percent of the ΔpopZ cells have ectopic poles, and many of these cells exhibit complex FDAA labeling patterns (Fig. 5H to L); however, in almost all cells, FDAA labeling occurs exclusively at poles or midcell regions. Together, these data suggest that PopZ is not required for polar growth, but the absence of PopZ allows growth to occur simultaneously from multiple poles.
FIG 5.
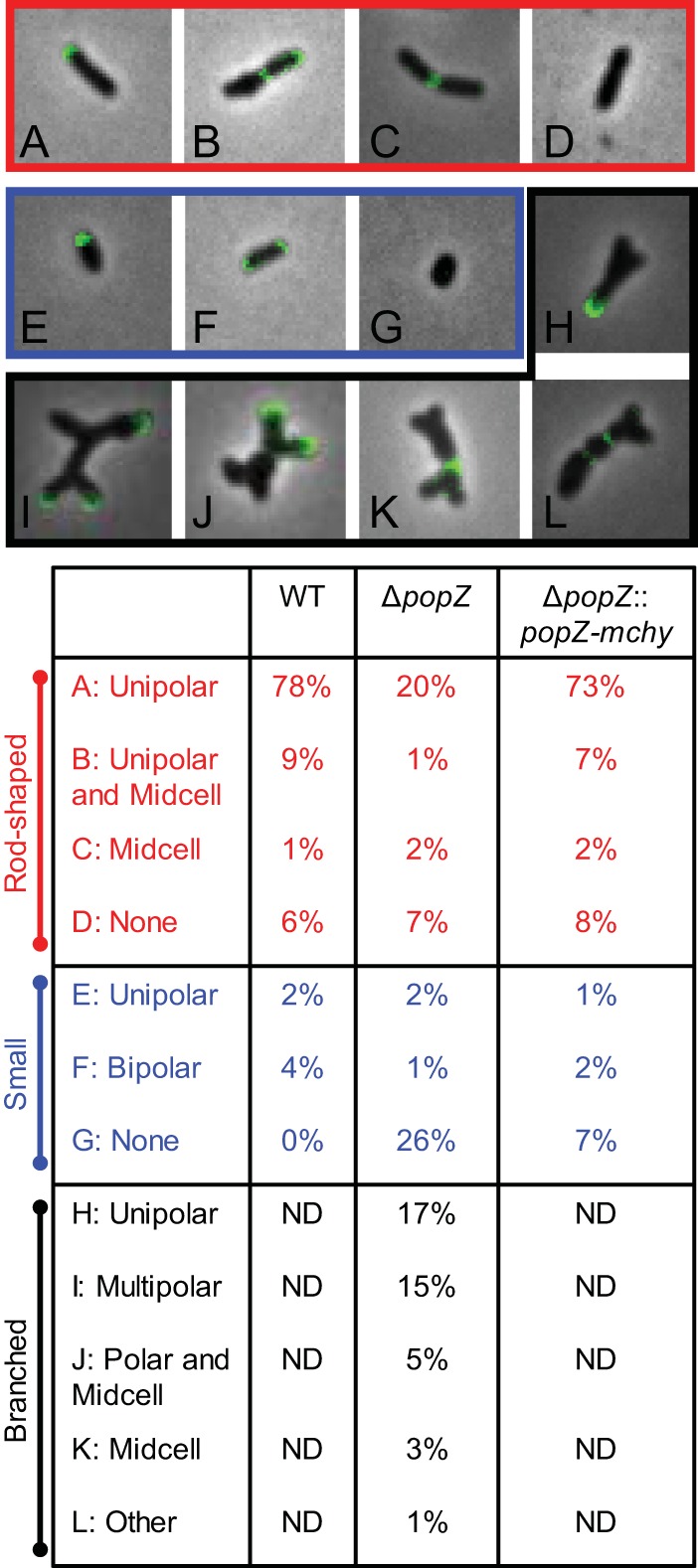
Deletion of popZ causes atypical growth patterning. Fluorescent d-amino-acid labeling was used as a proxy for sites of peptidoglycan synthesis in WT, ΔpopZ, and ΔpopZ::popZ-mchy strains. (Top) Representative images of cellular morphology and growth patterns from the ΔpopZ mutant are shown. Patterns observed in typical rod-shaped bacteria (red), small cells (<1.5 μm; blue), and branched cells (black) are shown and quantitated in the table below. One hundred fifty cells were labeled and categorized as shown in the table for each strain. Branched cells were not detected (ND) in WT and ΔpopZ::popZ-mchy strains.
Loss of PopZ causes minor changes in peptidoglycan composition.
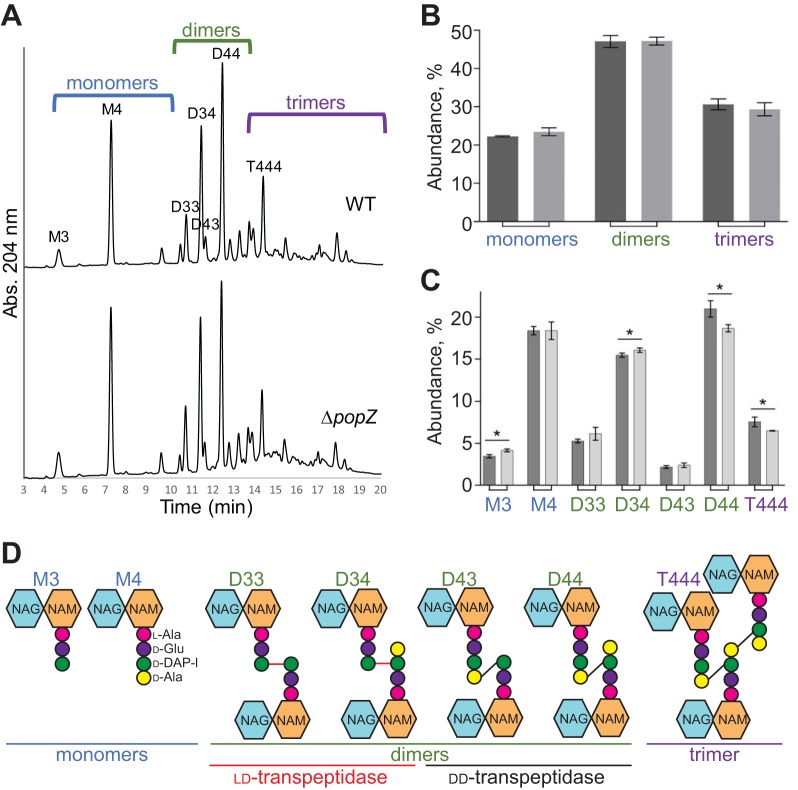
Although the FDAA labeling patterns were consistent with maintenance of proper cell wall biosynthesis at discrete subcellular localizations, we next determined if the absence of PopZ impacts peptidoglycan composition (Fig. 6A). Overall proportions of monomers (∼20% of the total muropeptides), dimers (∼50%), and trimers (30%) were not statistically different whether PopZ was present or absent (Fig. 6B). Further characterization of individual muropeptides does highlight a few subtle differences in the absence of PopZ, such as a decrease in muropeptides containing dd-cross-links (D43 and D44) and an increase in muropeptides with ld-cross-links (D33 and D34) (Fig. 6C and D). These changes indicate that PopZ may directly regulate the activity of a subset of cellular transpeptidases (i.e., dd-transpeptidase penicillin binding proteins [PBPs] or ld-transpeptidases). Alternatively, the altered cell morphology of the ΔpopZ cells, in particular the accumulation of ectopic poles, may be responsible for the differences in peptidoglycan composition.
FIG 6.
Peptidoglycan composition is slightly modified in ΔpopZ cells. (A) UPLC spectra of muropeptides derived from WT or ΔpopZ cells. Major muropeptides are labeled. M, monomers; D, dimers; T, trimers. Numbers indicate the length of the muropeptide stems and the position of cross-links in dimers and trimers. (B) Abundance of total monomers, dimers, and trimers in the muropeptide profile for WT (dark gray) and ΔpopZ (light gray) cells. (C) Quantitation of the major muropeptide peaks in WT (dark gray) and ΔpopZ (light gray) cells. For panels C and D, data shown are the average abundances of each muropeptide and are taken from analysis of three independent biological samples. Statistical significance is indicated with an asterisk. (D) Schematics of major muropeptides are shown. The monomers (blue), dimers (green), and trimer (purple) are labeled. Note that the type of cross-link is shown for each dimer and trimer. dd-Cross-links are shown in black, and ld-cross-links are shown in red.
Loss of PopZ results in lower frequency of FtsA-ring localization.
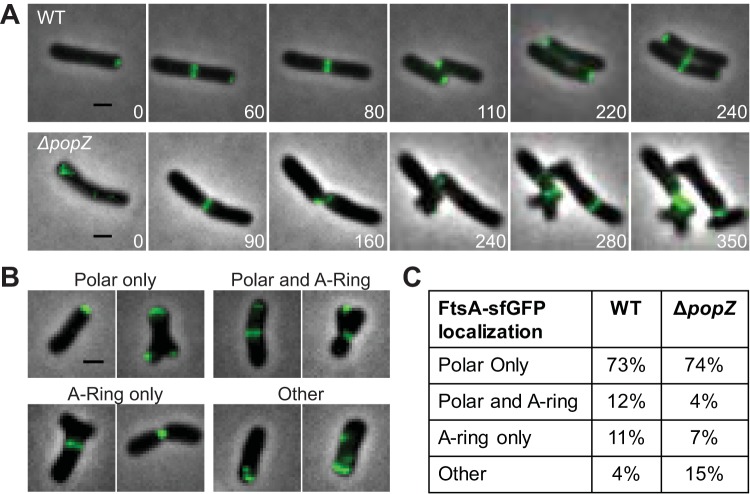
The accumulation of ectopic poles through sidewall branching and tip splitting, as well as the formation of small cells in the ΔpopZ strain, suggests that PopZ functions during the transition from polar peptidoglycan biosynthesis to midcell peptidoglycan synthesis to enable cell division. We next asked if the localization pattern of the essential cell division protein FtsA is impaired in ΔpopZ cells. In A. tumefaciens, FtsA localizes at the newly formed poles after cell division and remains at the growth poles during much of the elongation cycle before moving to midcell just prior to cell division (3, 6). We expressed ftsA-sfgfp under the control of its native promoter in both WT and ΔpopZ cells (Fig. 7). The FtsA localization pattern observed in WT cells was consistent with previous results (3, 6) (Fig. 7A, top). In the absence of PopZ, FtsA-sfGFP is consistently found at the growing pole, dissociates from these poles once growth is terminated, and forms rings as cells divide (Fig. 7A, bottom). In contrast to WT cells, we observe that in ΔpopZ cells, FtsA-rings do not form precisely at the midcell and the cells do not always divide where an FtsA-ring forms. When cell division failure leads to the formation of an ectopic pole, FtsA-sfGFP is present at the new growth pole. In order to better understand FtsA-sfGFP localization patterns in ΔpopZ cells, we observed the subcellular localization of FtsA-sfGFP in hundreds of individual cells (Fig. 7B and C). We find that the majority of WT and ΔpopZ cells have FtsA-GFP at poles, indicating that PopZ is not required for FtsA localization at the growth pole. We next quantified cells with FtsA-sfGFP rings rather than polar foci. In the WT population, 12% of cells undergo the transition from polar growth to midcell growth, as indicated by the presence of both polar and midcell FtsA-sfGFP. In contrast, only 4% of the ΔpopZ cells have both polar and midcell FtsA-sfGFP localization. Only 7% of ΔpopZ cells have FtsA-sfGFP present only as a ring, whereas FtsA-sfGFP rings were present in 11% of the WT population. Together, these data show that nearly a quarter of the WT population has an FtsA-ring, while only ∼10% of the ΔpopZ population has an FtsA-ring. In the ΔpopZ population, we observe an increase in transient localization of FtsA, often with multiple foci along the cell length (Fig. 7B and C, other). Altogether, these data show that PopZ is not required for formation of polar FtsA foci or ringlike structures; however, the mislocalization of FtsA and inefficiency of FtsA-ring formation suggest that PopZ plays a role in establishing the proper cell division site.
FIG 7.
FtsA forms polar foci and rings in ΔpopZ cells. (A) Time-lapse microscopy of FtsA-sfGFP in WT cells (top) shows a polar-to-midcell localization pattern. Time-lapse microscopy of FtsA-sfGFP in the ΔpopZ mutant (bottom) shows cells with unipolar foci, multipolar foci, and both stable and unstable ring structures. (B) Representative examples of FtsA-sfGFP localization patterns observed in ΔpopZ cells. All scale bars are 1 μm. (C) Percentage of cells with each category of FtsA-sfGFP localization pattern in WT (309 cells) and ΔpopZ (328 cells) strains.
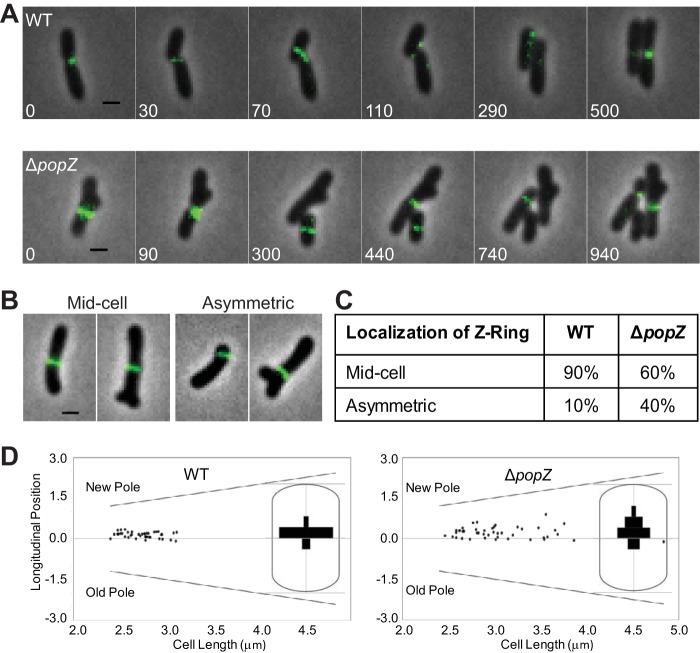
Loss of PopZ results in asymmetric Z-rings and constriction sites.
The reduction of FtsA-ring formation and production of minicells suggests that there is a cell division defect in the absence of PopZ. To better understand cell division in the ΔpopZ mutant, the localization pattern of the essential division protein FtsZ when fused to sfGFP (FtsZ-sfGFP) was observed (Fig. 8). Consistent with previous reports (3, 6), in WT cells FtsZ-sfGFP forms a constricting ring at the midcell in late predivisional cells (Fig. 8A, top). Following cell division, FtsZ-sfGFP foci are retained at the new pole for a portion of the cell cycle. As the cells elongate, FtsZ-sfGFP is released from the pole, exhibits transient localization, and then marks the new site of cell division. In ΔpopZ cells, FtsZ-sfGFP maintains its ability to form functional FtsZ-rings (Fig. 8A, bottom); however, the Z-rings formed are less consistent in size, placement, and stability. Time-lapse microscopy of ΔpopZ cells undergoing cell division reveals an unusually wide Z-ring (0 min) that marks the site of a cell division event. One of the resulting daughter cells is a short rod-shaped cell that eventually produces an FtsZ-ring at midcell (300 min); however, this cell does not elongate or divide, suggesting it lacks DNA, and the FtsZ-ring appears to disassemble (740 min). The other daughter cell is branched but forms a functional FtsZ-ring near midcell (440-min panel), producing two more daughter cells. These daughter cells form functional FtsZ-rings, although one is misplaced near the pole (740 min) and the other forms near midcell (940 min). To further characterize Z-ring placement, we analyzed the longitudinal position of the FtsZ-ring in individual cells (Fig. 8B and C). We considered FtsZ-rings to be positioned at midcell if they were located within 0.15 μm of the true midcell based on cell length measurements. Using this criterion, we observe that 90% of wild-type FtsZ-rings are found at midcell, whereas only 60% of FtsZ-rings are found at midcell in ΔpopZ cells (Fig. 8C). Among the 40% of ΔpopZ cells exhibiting asymmetric Z-rings, we observed that most of the Z-rings are found in close proximity to a pole (Fig. 8C). To determine if constriction sites are established near a particular pole, we oriented the cells by treating them with FM4-64, a fluorescent lipophilic dye which has been shown to preferentially label the old pole in A. tumefaciens (6). We were then able to map the longitudinal position of constriction sites in individual cells (Fig. 8D). In WT cells, constriction sites are identified in 41 of 330 cells analyzed and are located extremely close to the midcell (defined as a longitudinal position of 0). In contrast, constriction sites are observed in 41 of the 493 ΔpopZ cells analyzed and are more variable in position. Remarkably, there is a clear bias for asymmetric constriction sites to form closer to the new pole. Together, these data suggest that PopZ is not required for functional Z-ring formation; however, PopZ is required to ensure proper subcellular localization of the Z-ring.
FIG 8.
Analysis of FtsZ localization and constriction sites in ΔpopZ cells. (A, top) Time-lapse microscopy of FtsZ-sfGFP in WT cells shows dynamic localization of FtsZ foci before Z-ring formation at midcell. (Bottom) Time-lapse microscopy of FtsZ-sfGFP in ΔpopZ cells reveals atypical Z-ring size and positioning. (B) Representative images showing midcell and asymmetric Z-ring of ΔpopZ cells. All scale bars are 1 μm. (C) Quantification of Z-ring position in WT and ΔpopZ cells. Z-rings greater than 0.15 μm from midcell in either direction are defined as asymmetric. For this analysis, 77 Z-rings were analyzed in WT cells and 60 Z-rings were analyzed in ΔpopZ cells. (D) The longitudinal position of constriction sites is plotted against the cell length for 41 WT and 41 ΔpopZ cells. The position of each pole is shown with a diagonal line. A longitudinal position of zero is midcell. Positive values are closer to the new pole, whereas negative values are closer to the old pole. Insets show histograms illustrating the longitudinal position of constriction sites within a cell.
DISCUSSION
PopZ previously has been reported in C. crescentus to play a crucial role in chromosome segregation and the establishment of polar identity by serving as a landmark protein (8–10, 13–15, 20). The high degree of conservation between PopZCc and PopZAt in particular at the N terminus and C terminus (13) suggested that there is some conservation of PopZ function. Indeed, here we show the C terminus of PopZAt is important for proper subcellular localization, consistent with observations of PopZCc truncations, which demonstrate that the C terminus of PopZ is necessary and sufficient for polar localization (8, 9). Remarkably, PopCc and PopZAt exhibit striking differences in subcellular localization pattern (5). The localization of PopZ at the growing pole is reminiscent of other landmark proteins, such as DivIVA, which directs polar growth in Streptomyces (21, 22). Despite its strict localization to the growth pole, we find that PopZ is not required to direct polar peptidoglycan biosynthesis in A. tumefaciens. In the absence of PopZ, polar growth continues and peptidoglycan composition is only slightly modified, suggesting that PopZ is not required to maintain polar growth. How peptidoglycan biosynthesis is directed specifically to the growth pole during elongation remains an important and unanswered question.
Our observations suggest that PopZAt functions during the transition from polar peptidoglycan synthesis to midcell peptidoglycan synthesis as the cells divide. The deletion of popZ results in the formation of ectopic poles that arise from branching at sites of failed cell division events and growth tip splitting. In addition, we find that the loss of PopZ leads to mislocalization of two essential cell division proteins, FtsA and FtsZ, suggesting that PopZ directly or indirectly impacts the site of Z-ring formation, divisome assembly, and subsequent cell constriction. In the absence of PopZ, sites of constriction tend to form asymmetrically and are biased in positioning toward the new pole.
Why are FtsZ-rings positioned closer to the new pole in the absence of PopZ? Perhaps PopZ plays a direct role in regulating the localization of cell division proteins. Indeed, FtsZ and FtsA are retained at the growth pole following cell division and colocalize with PopZ during part of the cell cycle (5, 6); however, our data do not favor a direct role for PopZ in the localization of cell division proteins. In the absence of PopZ, polar foci of FtsA and FtsZ are observed, suggesting that PopZ is not required for the retention of cell division proteins at the pole following cell division. Furthermore, in most cells neither FtsA nor FtsZ becomes trapped at the pole in the absence of PopZ, suggesting that PopZ is not required for the release of cell division proteins from the growth pole. Finally, localization studies with photoconvertible mEos3.2-PopZ demonstrate that PopZ is transferred to the opposite pole following cell division (12), suggesting that PopZ is unlikely to participate in divisome assembly at midcell.
We favor an indirect role for PopZ in the regulation of divisome assembly. DAPI staining of ΔpopZ cells reveals that many cells lack DNA, consistent with a possible role for PopZ in chromosome segregation. Indeed, recent work from the Bowman laboratory has convincingly demonstrated that PopZAt is required for tethering the centromere of the circular chromosome at the new pole (12). One possible explanation for our observations is that A. tumefaciens uses a negative regulator of FtsZ assembly to coordinate chromosome segregation and cell division. For example, nucleoid occlusion proteins prevent FtsZ-ring assembly over chromosomes in other bacterial systems (23, 24). A simple explanation for our observations is that FtsZ-rings are assembling in the DNA-free zones formed due to the chromosome segregation defects in cells lacking PopZ; however, a nucleoid occlusion protein is not readily identifiable in the A. tumefaciens genome.
Alternatively, PopZ may function as a polar landmark protein and to directly or indirectly recruit negative regulators of FtsZ assembly to the pole. For example, MinCD proteins function as negative regulators for FtsZ assembly near the cell poles (25, 26), and minCDE genes are readily identifiable in the A. tumefaciens genome. PopZ may act as a polar targeting protein to recruit or stabilize MinCD at the new cell pole to prevent FtsZ-ring assembly near the growth pole. Since the Min system of A. tumefaciens remains uncharacterized, it is unclear if polar targeting proteins are necessary to establish a gradient of FtsZ-ring inhibition near the cell poles. Notably, saturating transposon mutagenesis suggests that the min genes are dispensable for cell survival in A. tumefaciens (27), and the absence of the entire Min system in the closely related bacterium Sinorhizobium meliloti does not impact cell growth or morphology (28), indicating that additional mechanisms to regulate the position of divisome assembly must exist.
We propose that PopZ has a conserved role in the coordination of chromosome segregation and cell division in alphaproteobacteria. In C. crescentus, ParB anchors the chromosome to the pole through its interaction with PopZ (8, 9) and regulates divisome assembly through its interactions with MipZ, a negative regulator of FtsZ-ring assembly (29, 30). Although A. tumefaciens does not have an MipZ homolog, our data suggest PopZ functions to directly or indirectly target regulators of FtsZ-ring assembly to the growth pole. Improved understanding of the mechanisms underlying FtsZ-ring positioning in A. tumefaciens will be crucial to understanding the precise role of PopZ in these processes.
In A. tumefaciens, PopZ marks the growth pole while PodJ marks the old pole (5). Remarkably, the phenotype described here for the ΔpopZ mutant is strikingly similar to the phenotype of a ΔpodJ mutant (11). Loss of either PopZ or PodJ causes ectopic pole formation, mislocalization of cell division proteins, and asymmetric cell division, suggesting that both proteins contribute to the regulation of divisome assembly. Furthermore, the absence of either PopZ or PodJ results in the production of nongrowing anucleate cells, suggesting that these proteins function in chromosome segregation. Both PopZ and PodJ have the capacity to act as polar landmark proteins and contribute to the new and old pole identity, respectively, by recruiting regulatory proteins to the poles. These findings highlight the need for further research to understand how PopZ and PodJ participate in the temporal and spatial coordination of cell cycle progression, peptidoglycan synthesis, and chromosome replication and segregation in A. tumefaciens.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
A list of all bacterial strains and plasmids used in this study is provided in Table S1 in the supplemental material. Agrobacterium tumefaciens C58C1 and derived strains were grown in ATGN minimal medium (31) without exogenous iron at 28°C with shaking. When appropriate, kanamycin was used at a working concentration of 300 μg/ml. When indicated, isopropyl-β-d-1-thio-galactopyranoside (IPTG) was used as an inducer at a concentration of 1 mM.
Construction of strains and plasmids.
A list of all strains and plasmids used in this study is provided in Table S1. To construct expression vectors for popZ-sfgfp and derived truncations, the respective coding sequence was amplified from genomic DNA, digested using restriction enzymes, and ligated into pSRKKm-Plac-sfgfp (32) using T4 ligase. Primers used for the construction of each popZ fragment are listed in Table S2.
To construct an expression vector for ftsA-sfgfp under the control of the native promoter, a gene block (IDT) (Table S2) that contains the qaz promoter and a multiple-cloning site was digested with EcoRI and HindIII and inserted into pSRKKm-Plac-sfgfp, creating pSRKKM-Pqaz-sfgfp. The ftsA coding sequence next was amplified using the primers indicated in Table S2 and inserted into pSRKKM-Pqaz-sfgfp to construct pSRKKM-Pqaz-ftsA-sfgfp.
To construct an ftsZ-sfgfp expression vector, we amplified the ftsZ coding sequence (Table S2) and inserted ftsZ into pSRKKm-Plac-sfgfp. ftsZ-sfgfp was not expressed well from the lac promoter. Thus, ftsZ-sfgfp was amplified from pSRKKm-Plac-ftsZ-sfgfp and inserted into pRVGFPC-2 (33) digested with NdeI and EcoRI to remove GFP. ftsZ-sfgfp amplification was digested and then ligated into the digested vector to create PRV-ftsZ-sfgfp. In A. tumefaciens, this vector provides a low level of constitutive expression of ftsZ.
The coding sequences for fliM and yfp were amplified from the A. tumefaciens genome or a source plasmid, respectively, using the primers indicated in Table S2. The two fragments were cloned into plasmid pSRKKM (34), cut with NdeI and HindIII by isothermal cloning (35). This plasmid (pGB1246) was transformed into Agrobacterium C58C1 strains with and without popZ (12).
All expression vectors were transformed into C58C1, C58C1ΔpopZ, and C58C1 ΔpopZ::mchy-popZ (12) strains, as indicated, using standard electroporation conditions (36).
Phase-contrast and fluorescence microscopy.
A small volume (∼0.8 μl) of cells in exponential phase (optical density at 600 nm [OD600] of 0.4 to 0.6) was applied to a 1% agarose pad as described previously (1). Phase-contrast and epifluorescence microscopies were performed with an inverted Nikon Eclipse TiE and a QImaging Rolera em-c2 1K electron-multiplying charge-coupled device (EMCCD) camera with Nikon Elements Imaging software. To visualize DNA, cells were grown in ATGN medium to exponential phase and ethanol fixed. Cells were resuspended in phosphate-buffered saline (PBS) containing 1 μg/ml of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) for 5 min. Excess dye was washed out with PBS and cells were imaged.
Quantification of cell length distributions.
Cells were grown overnight in ATGN medium. Cells were diluted in ATGN to an OD600 of 0.10 and allowed to grow until reaching an OD600 of 0.4 to 0.6. Live cells were imaged using phase-contrast microscopy, and cell length distributions of at least 800 cells per strain were determined using the longest medial axis as measured by MicrobeJ software (37).
Localization of PopZ-GFP truncations.
Cells were grown overnight in ATGN medium in the presence of 1 mM IPTG and 300 μg/ml kanamycin. Cells were diluted to an OD600 of 0.10 and allowed to grow until reaching an OD600 of 0.4 to 0.6. Live cells were then placed onto a 1.25% agarose-ATGN pad, and phase-contrast and epifluorescence microscopies were performed.
UPP labeling.
Wheat germ agglutinin lectin conjugated to Alexa Fluor 488 (WGA-AF488) (Life Technologies) was used to detect polar polysaccharide in A. tumefaciens. In all experiments, a concentration of 0.5 μg/ml of WGA-AF488 was used, and exponential-phase cells were incubated for 30 min before excess lectin was removed by washing. One microliter of washed culture was spotted on an agarose pad and cells were imaged.
Short-term binding assay.
The short-term binding assay was conducted as described previously (38), with slight modifications. Briefly, bacteria were grown to exponential phase, and 2 ml of culture at an OD600 of 0.4 was placed in each well of a 6-well plate containing a glass coverslip and incubated for 1 h. Planktonic cells were removed and coverslips were washed by pipetting media over the surface of the coverslip at least 4 times. Excess liquid was wicked off from the coverslip, and the coverslip was inverted onto a microscope slide and immediately imaged.
Biofilm assay.
Biofilm assays were conducted as described previously (38). Briefly, bacteria were grown to mid-log phase in ATGN medium and diluted to an OD600 of 0.03. Three milliliters of diluted bacteria was placed in the wells of 12-well polystyrene plates containing polyvinyl chloride coverslips placed vertically in each well. Plates were incubated at room temperature for 48 h. After incubation, the OD600 of cultures from each well was measured to normalize for the density of cell culture. Coverslips were then removed and rinsed with distilled H2O to remove planktonic cells. Coverslips were stained with 0.1% crystal violet for 5 to 10 min and destained with 33% acetic acid. The A600 of the solubilized crystal violet was measured. The biofilm score was calculated as the A600/OD600 ratio to allow for normalization based on the cell density of each strain. Two biological replicates were completed in triplicate for each experiment.
FliM-YFP microscopy and image analysis.
Cells were grown to exponential phase in ATGN and induced with 400 μM IPTG for 2 to 3 h prior to time-lapse observation by fluorescence microscopy (as described in reference 20) on ATGN-agarose pads. To determine the rate of cell growth and the movement of FliM-YFP foci from the nearest cell pole, the measuring tool in Zen software (Zeiss) was used to make length measurements at each time point. Measuring every available interval from 34 different cells from two independent experiments yielded a total number of 72 intervals.
Quantification of cell morphologies and FDAA labeling patterns.
Cells were grown overnight in ATGN medium, diluted under the same conditions to an OD600 of 0.10, and allowed to grow until reaching an OD600 of 0.4 to 0.6. At this point cells were labeled with the fluorescent d-amino acid (FDAA) HCC amino-d-alanine (HADA) as previously described (2). Immediately following labeling, cells were ethanol fixed to prevent further growth. Phase-contrast and epifluorescence microscopies were performed. One hundred fifty cells for each strain were used to quantify the three morphological categories, normal (>1.5 μm without branches), small (<1.5 μm), and branched (containing 3 or more poles), using the ImageJ cell counter plug-in. These cells were further subdivided based on differential FDAA labeling among the three morphologies.
Peptidoglycan compositional analysis.
Cell cultures were grown overnight in 3-ml culture tubes of ATGN minimal medium at 28°C with shaking. The 3-ml cultures were then added to 50-ml flasks of fresh ATGN and allowed to grow under the same conditions until reaching an exponential-phase OD600 of 0.5 to 0.6. Cultures were then pelleted by centrifugation at 4,000 × g for 15 min. Pellets were resuspended in a solution of 3 ml ATGN and 6 ml of 6% SDS. Cells were boiled while being simultaneously stirred by magnetic bars for 4 h. After 4 h, boiling was halted but agitation was continued overnight. Peptidoglycan was pelleted by centrifugation for 13 min at 60,000 rpm (TLA100.3 Beckman rotor, Optima Max-TL ultracentrifuge; Beckman), and the pellets were washed 3 to 4 times by repeated cycles of centrifugation and resuspension in water. The pellet from the final wash was resuspended in 50 μl of 50 mM sodium phosphate buffer, pH 4.9, and subjected to overnight digestion with 100 μg/ml of muramidase at 37°C. Muramidase digestion was stopped by boiling for 4 min. Coagulated protein was removed by centrifugation for 15 min at 15,000 rpm in a desktop microcentrifuge. The muropeptides were mixed with 15 μl 0.5 M sodium borate and subjected to reduction of muramic acid residues into muramitol by sodium borohydride (10 mg/ml final concentration, 20 min at room temperature) treatment. Samples were adjusted to pH 3 to 4 with orthophosphoric acid and filtered (0.2-μm filters).
Analysis of muropeptides was performed on an Acquity ultraperformance liquid chromatography (UPLC) BEH C18 column (130 Å, 1.7 μm, 2.1 mm by 150 mm; Waters, USA) and detected at A204 with an Acquity UPLC UV-visible detector. Muropeptides were separated using a linear gradient from buffer A (50 mM sodium phosphate buffer, pH 4.35) to buffer B (50 mM sodium phosphate buffer, pH 4.95, 15% [vol/vol] methanol) with a flow of 0.25 ml/min in a 20-min run. Individual muropeptides were quantified from their integrated areas using samples of known concentration as standards. Muropeptide abundance was statistically compared using unpaired t test.
Distribution of constriction sites.
Exponential-phase cells were first labeled with the lipophilic dye FM4-64, which preferentially labels the old pole (6). Fifteen fields of cells were imaged with phase-contrast and epifluorescence microscopies. Cells were detected and the positions of constrictions were determined using MicrobeJ (37). Cells were orientated by defining the old pole as the pole with the most intense FM4-64 labeling. Potential constriction sites were identified using a local maximum algorithm to identify the lowest width value along the width profile excluding both ends of the medial axis (37). For each potential constriction, the ratio of the median cell width/minimal cell width is calculated. Thus, larger ratio values represent deeper constrictions. A threshold of a 0.44 ratio value was used to identify deep constrictions. The longitudinal position of the constriction site was plotted against the cell length. A value of 0 denotes midcell, negative numbers represent positions closer to the old pole, and positive numbers represent positions closer to the new pole. Additionally, histograms plotting the relative longitudinal position of the septum were plotted on a cell outline.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Electron Microscopy Core at the University of Missouri for assistance collecting the transmission electron micrographs. We are grateful to George Smith, Chiqian Zhang, and members of the Brown laboratory for helpful discussions and critical reading of the manuscript.
P.J.B.B. and M.H. were supported by the National Science Foundation, IOS1557806. G.R.B. and A.K.S. were supported by the National Science Foundation, MCB-1518171, and by an Institutional Development Award (IDeA) from the National Institutes of Health, 2P20GM103432. F.C. and A.A. receive funding support from the Laboratory for Molecular Infection Medicine Sweden, the Knut and Alice Wallenberg Foundation, Kempe, and the Swedish Research Council. A.A. is supported by a MIMS/VR Ph.D. position.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/JB.00111-17.
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00101-17.
REFERENCES
- 1.Brown PJB, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A 109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuru E, Velocity Hughes H, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl 51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameron TA, Anderson-Furgeson J, Zupan JR, Zik JJ, Zambryski PC. 2014. Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio 5:e01219-14. doi: 10.1128/mBio.01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margolin W. 2009. Sculpting the bacterial cell. Curr Biol 19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grangeon R, Zupan JR, Anderson-Furgeson J, Zambryski PC. 2015. PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 112:11666–11671. doi: 10.1073/pnas.1515544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zupan JR, Cameron TA, Anderson-Furgeson J, Zambryski PC. 2013. Dynamic FtsA and FtsZ localization and outer membrane alterations during polar growth and cell division in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 110:9060–9065. doi: 10.1073/pnas.1307241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treuner-Lange A, Sogaard-Andersen L. 2014. Regulation of cell polarity in bacteria. J Cell Biol 206:7–17. doi: 10.1083/jcb.201403136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong SH, Jones Y, Lee JH, Downing KH, Ellisman MH, McAdams HH, Shapiro L. 2010. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol 76:173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson-Furgeson JC, Zupan JR, Grangeon R, Zambryski PC. 2016. Loss of PodJ in Agrobacterium tumefaciens leads to ectopic polar growth, branching, and reduced cell division. J Bacteriol 198:1883–1891. doi: 10.1128/JB.00198-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrle H, Guidry J, Iacovetto R, Salisbury A, Sandidge D, Bowman GR. 2017. The polar organizing protein PopZ is required for chromosome segregation in Agrobacterium. J Bacteriol 199:e00111-17. doi: 10.1128/JB.00111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laloux G, Jacobs-Wagner C. 2013. Spatiotemporal control of PopZ localization through cell cycle-coupled multimerization. J Cell Biol 201:827–841. doi: 10.1083/jcb.201303036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowman GR, Perez AM, Ptacin JL, Ighodaro E, Folta-Stogniew E, Comolli LR, Shapiro L. 2013. Oligomerization and higher-order assembly contribute to sub-cellular localization of a bacterial scaffold. Mol Microbiol 90:776–795. doi: 10.1111/mmi.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ptacin JL, Gahlmann A, Bowman GR, Perez AM, von Diezmann AR, Eckart MR, Moerner WE, Shapiro L. 2014. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci U S A 111:E2046–E2055. doi: 10.1073/pnas.1405188111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heindl JE, Wang Y, Heckel BC, Mohari B, Feirer N, Fuqua C. 2014. Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium. Front Plant Sci 5:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell M, Brown PJB. 2016. Building the bacterial cell wall at the pole. Curr Opin Microbiol 34:53–59. doi: 10.1016/j.mib.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol 89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt PM, Danhorn T, Fuqua C. 2007. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J Bacteriol 189:8005–8014. doi: 10.1128/JB.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes JA, Follett SE, Wang H, Meadows CP, Varga K, Bowman GR. 2016. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc Natl Acad Sci U S A 113:12490–12495. doi: 10.1073/pnas.1602380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flardh K, Richards DM, Hempel AM, Howard M, Buttner MJ. 2012. Regulation of apical growth and hyphal branching in Streptomyces. Curr Opin Microbiol 15:737–743. doi: 10.1016/j.mib.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Hempel AM, Wang SB, Letek M, Gil JA, Flardh K. 2008. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol 190:7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernhardt TG, de Boer PA. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu LJ, Errington J. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Rowlett VW, Margolin W. 2013. The bacterial Min system. Curr Biol 23:R553–R556. doi: 10.1016/j.cub.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 26.Rowlett VW, Margolin W. 2015. The Min system and other nucleoid-independent regulators of Z ring positioning. Front Microbiol 6:478. doi: 10.3389/fmicb.2015.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtis PD, Brun YV. 2014. Identification of essential alphaproteobacterial genes reveals operational variability in conserved developmental and cell cycle systems. Mol Microbiol 93:713–735. doi: 10.1111/mmi.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng J, Sibley CD, Zaheer R, Finan TM. 2007. A Sinorhizobium meliloti minE mutant has an altered morphology and exhibits defects in legume symbiosis. Microbiology 153:375–387. doi: 10.1099/mic.0.2006/001362-0. [DOI] [PubMed] [Google Scholar]
- 29.Kiekebusch D, Michie KA, Essen LO, Lowe J, Thanbichler M. 2012. Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol Cell 46:245–259. doi: 10.1016/j.molcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thanbichler M, Shapiro L. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 31.Morton ER, Fuqua C. 2012. Laboratory maintenance of Agrobacterium. Curr Protoc Microbiol 24:3D.1–3D.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueroa-Cuilan W, Daniel JJ, Howell M, Sulaiman A, Brown PJ. 2016. Mini-Tn7 insertion in an artificial attTn7 site enables depletion of the essential master regulator CtrA in the phytopathogen Agrobacterium tumefaciens. Appl Environ Microbiol 82:5015–5025. doi: 10.1128/AEM.01392-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res 35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan SR, Gaines J, Roop RM II, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 36.Morton ER, Fuqua C. 2012. Genetic manipulation of Agrobacterium. Curr Protoc Microbiol 25:3D.2.1–3D.2.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ducret A, Quardokus EM, Brun YV. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morton ER, Fuqua C. 2012. Phenotypic analyses of Agrobacterium. Curr Protoc Microbiol 25:3D.3.1–3D.3.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.