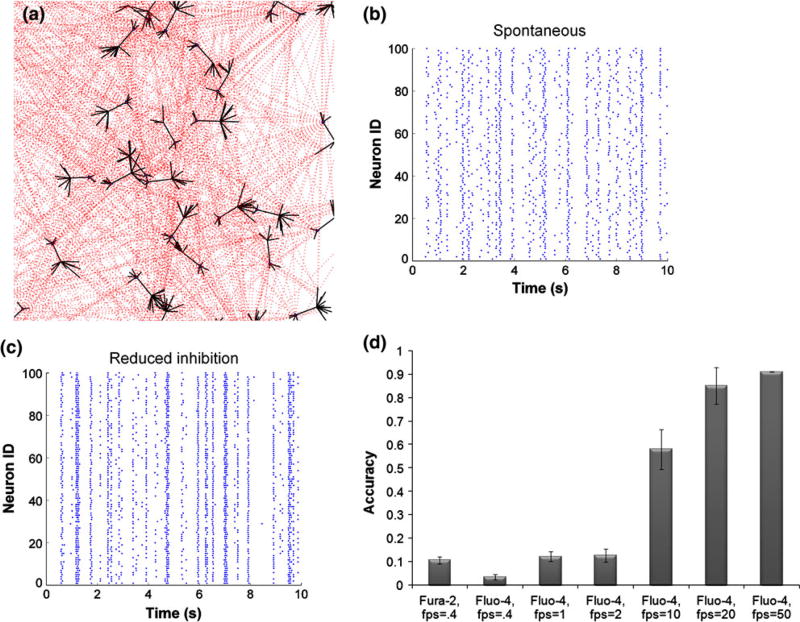
Figure 1. In silico model of neural network activity.
(a) Pyramidal excitatory neuronal network was constructed, where each neuron connected to 20 other neurons on average, chosen from a Poisson distribution. Synaptic connections occurred on dendritic spines, populated with AMPA and NMDA receptors. (b, c) Stochastic integrate-and-fire model reproduced spontaneous activity patterns, composed of flickering events, intermixed with network-wide bursts. Reducing the strength of the inhibitory network increased synchronous oscillations. (d) Calcium transients predicted from underlying action potentials were converted to fluorescence traces for fura-2 and fluo-4, taking into account their unique buffering properties. The accuracy of resolving action potentials using fluorescent dye is greater for fluo-4 and increases with imaging acquisition speed.