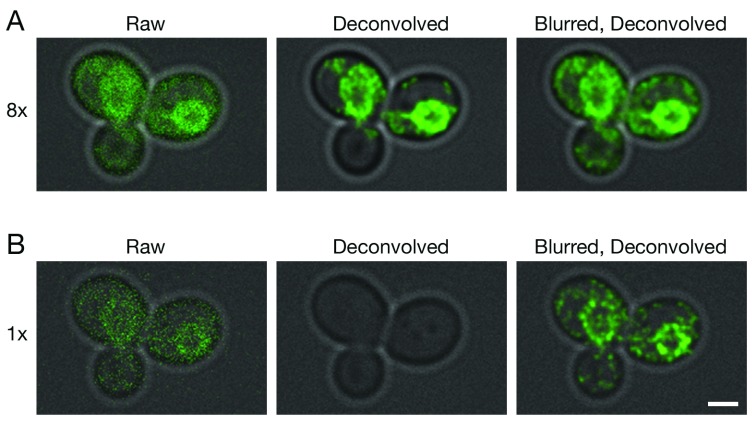
Figure 2. Improved deconvolution of non-punctate fluorescence signals with a Gaussian blur prefilter.
Gene replacement in Saccharomyces cerevisiae was used to label ER membranes with Hmg1-GFP ( Koning et al., 1996). A confocal Z-stack was captured with line accumulation set to ( A) 8x or ( B) 1x. The data were average projected either with no processing, or after deconvolution with Huygens, or after prefiltering with a 2D Gaussian blur using a radius of 0.75 pixels followed by deconvolution. Fluorescence data are superimposed on differential interference contrast images of the cells (gray). Scale bar, 2 µm.