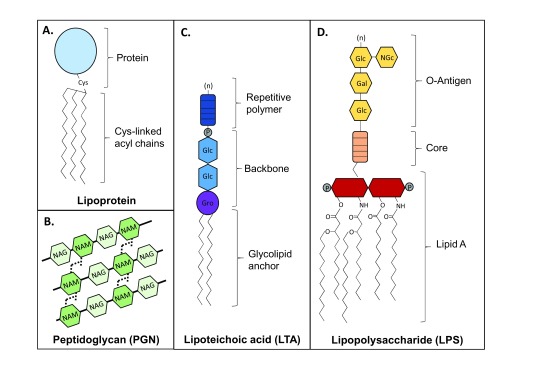
Figure 2. Structural variety of microbial lipid pathogen-associated molecular patterns.
( A) Lipoproteins are anchored to the bacterial membrane by cysteine-linked acyl chains and have been described in both Gram-positive and Gram-negative bacteria. ( B) Peptidoglycan (PGN) consists of a sugar polymer composed of β-(1,4)-linked N-acetylglucosamine (NAG) and N-acetyl muramic acid (NAM). Peptide chains of three to five amino acids are attached to the NAM residues and cross-linked to a chain from another sugar polymer strand, creating a mesh-like structure. ( C) Lipoteichoic acid (LTA) is a Gram-positive surface polymer abundant in the bacterial envelope. The LTA structure can vary and is classified into categories by characteristics such as its backbone type and repetitive polymer composition (types I–V). Many LTAs contain a glycerophosphate (GroP) backbone structure attached to a glycolipid anchor. The repeating unit (marked with “n”) can also vary and may contain glycerol, glycose, galactose, ribitol, phosphate, and other related derivatives. Type I/II LTA backbone is represented here. ( D) Lipopolysaccharide (LPS) is a major component of the outer leaflet of the outer membrane of Gram-negative bacteria. The hydrophobic membrane anchor, lipid A, can vary in acyl chain length and number and can be mono- or bis-phosphorylated. The lipid A molecule is responsible for innate immune recognition by TLR4. Lipid A is connected to a hydrophilic core polysaccharide chain, followed by a repeating (marked with “n”) oligosaccharide chain (the O-antigen), which is specific to the bacterial serotype.