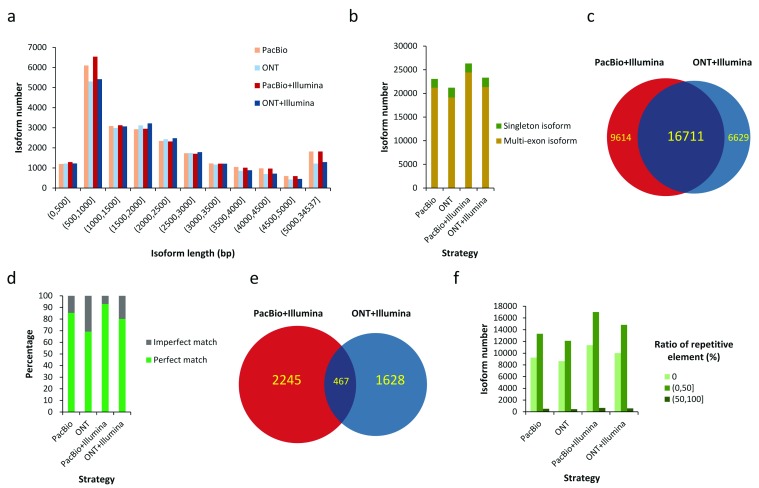
Figure 4. Isoform identification in human embryonic stem cells.
( a) Length distribution of isoforms identified by full-length by long read only and Hybrid-Seq strategies. ( b) Numbers of identified isoforms with single exon (singleton isoform) and multiple exons (multi-exon isoform). ( c) Overlap between isoforms identified by two Hybrid-Seq strategies. ( d) Accuracy of splice sites detected by four strategies. Perfect means the detected splice sites exactly match known splice sites annotated by Gencode (version 24). Imperfect means the detected splice sites are shorter or longer than known splice sites annotated by Gencode (version 24). ( e) Overlap between novel isoforms identified by two Hybrid-Seq strategies. ( f) Numbers of identified isoforms with different ratios of repetitive elements. ONT: Oxford Nanopore Technologies; PacBio: Pacific Biosciences.