Abstract
Aim:
Subclinical mastitis in bovines is mainly responsible for the huge economic loss of the dairy farmers, of which Pseudomonas aeruginosa is one of the causative agents. The study was aimed at a screening of suspected milk samples from different cattle farms of West Bengal for detection and confirmation of P. aeruginosa strains followed by their characterization.
Materials and Methods:
Around 422 milk samples were screened from different dairy farms primarily by on-spot bromothymol blue (BTB) test and then in the lab by somatic cell counts (SCC) to finally consider 352 samples for detection of P. aeruginosa. Selective isolation and confirmation of the isolates were done using selective media, viz., cetrimide and Pseudomonas agar followed by confirmation by fluorescent technique. Molecular characterization of the strains was done by polymerase chain reaction for the presence of toxA (enterotoxin A, 352 bp) and exoS (exoenzyme S, 504 bp) genes.
Results:
Approximately, 371 (87.9%) samples were positive in on-spot BTB test among which 352 (94.8%) samples revealed high SCC values (more than 3 lakh cells/ml) showing infection when screened. Among these, 23 (6.5%) samples yielded typical Pseudomonas sp. isolates out of which only 19 (5.4%) isolates were confirmed to be P. aeruginosa which showed characteristic blue-green fluorescence due to the presence of pigment pyoverdin under ultraviolet light. Out of these 19 isolates, 11 isolates were positive for toxA, 6 isolates for exoS, and 2 for both these pathogenic genes.
Conclusion:
Approximately, 5.4% cases of bovine subclinical mastitis infections in South Bengal were associated with P. aeruginosa which possess pathogenic genes such as toxA (63.2%) and exoS (36.8%).
Keywords: bovines, characterization, exoS, Pseudomonas aeruginosa, subclinical mastitis, toxA
Introduction
India is one among the largest milk producing countries in the world right now. Dairy industry plays a significant role in livestock economy by generating self-employment. Dairy industry by cooperative society brought socioeconomic transformation in innumerable small, marginal dairy farmers and downtrodden people of mainly western part of India along with other parts the country. These livestock farmers in India mainly contribute the major total national milk production which now rose up to approximately 132.4 million tons in 2012-13 from 17 million tons in 1950-51 [1]. One of the major drawbacks in this industry is a loss of milk production due to mastitis and mastitis related problems of the herd. Subclinical mastitis is one of the major causes of milk production loss in India [2]. An estimated total loss of INR 26460 million due to subclinical mastitis of cows in India was reported by Dua [3]. As it is very difficult diagnose in field level by cattle owners, this may lead to chronic infection resulting into irreparable damage of mammary gland of infected cattle, thus culling or elimination cost is also involved in the list of economic losses. Approximately, 12% loss in milk yield due to subclinical mastitis with an average loss of Rs. 1016/- per subclinical mastitis case was also reported by Patel et al. [4]
Studies on the etiological factors of subclinical mastitis revealed that bacteria were the major cause followed by different types of viruses, algae, and mycoplasma [2]. One of the major bacterial pathogens associated with this infection Pseudomonas aeruginosa [5]. A study at Jammu and Kashmir, India [5] revealed high positivity of subclinical mastitis in a herd of crossbred cows caused by different pathogens along with P. aeruginosa (3.6%). These Gram-negative bacteria may possess several pathogenic factors responsible for their pathogenicity, among which exotoxin A (toxA) and exoenzyme S (exoS) are two major fatal weapons which remain associated with subclinical mastitis infection [6] in bovines.
Therefore, this study was aimed at the detection and molecular characterization of P. aeruginosa from subclinical mastitis cases of bovines from different districts of West Bengal for developing an idea about the significance of this infection.
Materials and Methods
Ethical approval
The study was approved by Institutional Biosafety Committee of the University and as per the Committee for the Purpose of Control and Supervision on Experiments on Animals rules; it does not require any approval of Institutional Animal Ethics Committee.
Sample collection
All milk samples (422 in no.) were screened by on-spot positive bromothymol blue (BTB) test according to the standard method [7] with BTB paper, and 371 samples were collected (aseptically directly from the teats in sterile vials) from different organized government farms (8 farms were covered) and small rural farmers (46 farmers were considered for collection) of different districts of South Bengal for this study during October 2013-April 2014 (i.e., total 7 months which were the time period fixed for this study). The samples were collected from cattle irrespective of age and species, and no specific antibiotics were used as per farm records. All 371 samples were tested somatic cell counts (SCC) study in the lab as per the method described by Sharma et al. [8] with slight modification.
Dried smears of suspected milk samples were stained by Newman’s modified stain (HiMedia) followed by counting of somatic cells and tentatively infected samples (n=352) with SCC value above 300,000/ml [9] were collected finally (Table-1) and stored at 4°C for further study.
Table-1.
Distribution pattern of positive samples in different farms/regions.
| Name of the regions | Total collected samples | BTB positive (%) | SCC positive samples with over 3 lakh cells/ml (%) | Average SCC value in lakh cells/ml |
|---|---|---|---|---|
| Haringhata cattle farms | 56 | 46 (82.1) | 43 (93.5) | 3.26 |
| Kalyani SLFs | 49 | 41 (83.7) | 37 (90.2) | 3.54 |
| Borsul area | 98 | 86 (87.7) | 81 (94.2) | 4.88 |
| Memari area | 106 | 100 (94.3) | 97 (97.0) | 3.92 |
| Mogra area | 113 | 98 (86.7) | 94 (95.9) | 4.67 |
| Total | 422 | 371 (87.9) | 352 (94.8) | 4.05 |
SCC=Somatic cell counts, BTB=Bromothymol blue
Isolation and characterization
Tentatively subclinical mastitis infected samples after collection were cultured for detection of desired pathogen. Each milk sample under study was streaked on sterile nutrient agar plates after overnight enrichment followed by overnight incubation at 37°C. Bacterial colonies with green color pigmentation were examined morphologically by Gram’s staining method and all Gram-negative bacilli found were further cultured on sterile cetrimide agar plates for final purification [10]. The greenish single colonies were picked up in sterile cetrimide agar slants for further characterization by different biochemical tests, viz., such as oxidase test, catalase test, nitrate reduction, indole test, methyl red test, Voges-Proskauer test, citrate utilization and glucose fermentation test as per Quinn et al. [10] and Carter and Wise [11]. All positive samples were tested twice to be sure of the result.
Confirmation of P. aeruginosa strains
Production of pigments such as pyocyanin and pyoverdin by the purified isolates were tested by “fluorescent technique” after growing the isolates on cetrimide and Pseudomonas agar followed by ultraviolet (UV) illumination [12] for confirmation.
Detection of pathogenic genes in P. aeruginosa isolates
Pathogenic P. aeruginosa isolates were confirmed by detection for enterotoxin A (toxA) and exoenzyme S (exoS) genes in them. DNA isolation and detection of pathogenic genes in all positive isolates were performed following methods of Lanotte et al. [13] and Brenner et al. [14]: Young broth culture (at 10 ml) of each isolate was harvested in 15 ml TE buffer (40 mM Tris/HCl, 2 mM EDTA, pH 8.0) and lysed in 220 µl of a 25% (w/v) aqueous solution of sodium dodecyl sulfate (SDS) and 30 µl pronase (an enzyme used for degradation of proteins during isolation of DNA). The mixture was then incubated overnight at 37°C to allow cell lysis. DNA was extracted [14] and was resuspended in 1 ml 1× TE for further use. A standard culture of P. aeruginosa (ATCC No. 27853) (as positive control) and one Escherichia coli isolate (departmental isolate) (as negative control) were used in this study for confirmation.
Two sets of primers (for toxA (352bp) (FP 5’GGTAACCAGCTCAGCCACAT3’, RP 5’ TGATG TCCAGGTCATGCTTC3’) and exoS (504bp) genes (FP 5’CTTGAAGGGACTCGACAAGG3’, RP 5’TTCAGGTCCGCGTAGTGAAT3’)) as per Lanotte et al. [13] were utilized to amplify the specific gene in thermal cycler under specific polymerase chain reaction (PCR) conditions as follows.
The PCR mixture contained PCR buffer (10 mM Tris/HCl, 50 mM KCl, 1.5 mM MgCl2, pH 8.3), 200 µM of each dNTP, 12.5 pmol of each primer, dimethyl sulfoxide at a final concentration of 4%, 1U Ampli Taq DNA polymerase and 25 ng DNA template (protocol followed as described by Lanotte et al.) [13]. The DNA was amplified in a thermal cycler (Eppendorf, Germany) using the following protocol: 94°C for 3 min, 30 cycles of 94°C for 30 s, 55°C for 1 min and 72°C for 1 min 30 s, and 72°C for 5 min. Each gene was amplified separately. PCR products were separated in a 1% agarose gel for 1 h at 100 V, stained with ethidium bromide and detected by UV transillumination.
Statistical analysis
All data recorded in this study were statistically analyzed using general linear model of IBM SPSS software (version 20).
Results
Primary screening of all milk samples by on-spot BTB test revealed that approximately 371 (87.9%) samples were positive out of which samples from Memari (94.3%) and Borsul (87.7%) blocks were highly positive than other farms. The somatic cell study of these samples showed that almost all (94-97%) samples were having high SCC average values of 3.92-4.88 lakh cells/ml (Table-1) which were indicative of infection. Samples with lower SCC values or normal SCC values were not considered for bacterial isolation.
On testing for bacterial isolation, a total of 23 (6.5%) samples were found to be positive for Pseudomonas sp. as they showed characteristic bluish-green pigmentation on cetrimide agar and were found to be Gram-negative bacilli in nature (Figure-1). Biochemical characterization of these isolates revealed that 21 samples were typical in nature, i.e., positive for oxidase, catalase, citrate utilization, nitrate reduction, and glucose fermentation whereas were negative for methyl red, Voges-Proskauer, and indole tests. Two [2] Pseudomonas sp. samples showed variable result in glucose fermentation test as revealed in this study.
Figure-1.
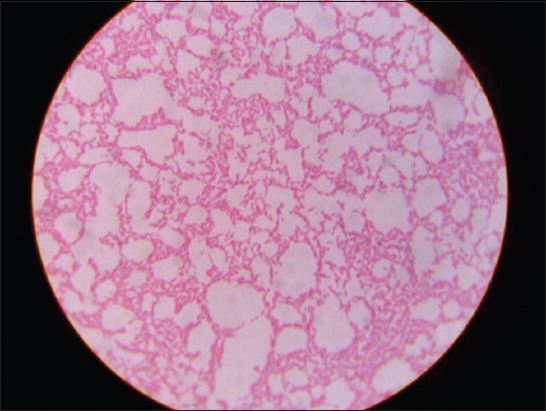
Pink colored bacilli of Pseudomonas aeruginosa isolates in Gram’s staining.
All 23 samples positive for Pseudomonas sp. (including 2 variables) were tested by fluorescent technique to detect 19 (5.4%) isolates showing characteristic blue-green fluorescence due to pigment pyoverdin under UV light (Figure-2) which is confirmatory for P. aeruginosa. Samples from Memari block showed highest positivity (7.2%) followed by others (Table-2). Molecular characterization of all 19 positive isolates revealed that 11 isolates to be positive for the presence of toxA (352 bp) gene, 6 isolates for exoS (504 bp) gene, and 2 isolates to possess both the genes (Table-2, Figures-3 and 4). The presence of toxA (63.2%) was found to be much higher than that of exoS (36.8%) in P. aeruginosa strains as found in thus study.
Figure-2.
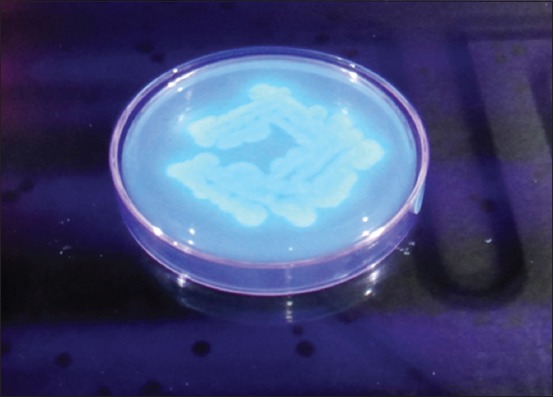
Blue-green fluorescence under ultraviolet light of Pseudomonas aeruginosa isolates.
Table-2.
Distribution pattern of Pseudomonas aeruginosa isolates in different regions with genetic characterization.
| Parameters of characterization | Name of the regions | |||||
|---|---|---|---|---|---|---|
| Haringhata cattle farms | Kalyani SLFs | Borsul block | Memari area | Mogra area | Total | |
| SCC positive samples | 43 | 37 | 81 | 97 | 94 | 352 |
| Positive Pseudomonas sp. detected | 1 | 2 | 6 | 8 | 6 | 23 |
| Positive Pseudomonas aeruginosa confirmed | 1 | 2 | 4 | 7 | 5 | 19 |
| Percentage of positivity | 2.3 | 5.4 | 4.9 | 7.2 | 5.3 | 5.4 |
| Pseudomonas aeruginosa with toxA | 1 | 1 | 2 | 3 | 4 | 11 |
| Pseudomonas aeruginosa with exoS | 0 | 1 | 2 | 3 | 0 | 6 |
| Pseudomonas aeruginosa with both exoS+toxA | 0 | 0 | 0 | 1 | 1 | 2 |
SCC=Somatic cell counts
Figure-3.
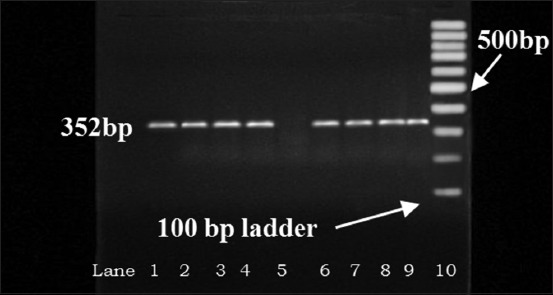
Detection of toxA gene (352 bp) in Pseudomonas aeruginosa isolates by polymerase chain reaction. Lane 2-4, 6-9: Test samples, 1: Positive control, 5: Negative control.
Figure-4.
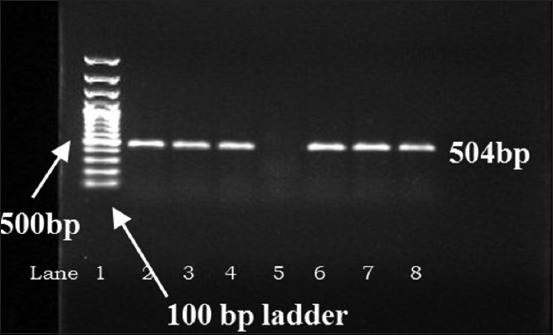
Detection of exoS gene (504 bp) in Pseudomonas aeruginosa isolates by polymerase chain reaction. Lane 3-4, 6-8: Test samples, 2: Positive control, 5: Negative control.
Discussion
Approximately, 371 (87.9%) samples were found to positive by on-spot BTB test (which indicates tentative infection) in this study which is more or less matching with the reports of Harini and Sumathi [15], Marschke and Kitchen [16] and Singh et al. [17] who also detected approximately 75-90% positivity in their study for subclinical mastitis in bovines. The change in pH of the infected milk samples can be detected by this BTB test, and as the change may be due to other noninfective causes too, that’s why secondary screening with SCC study was performed and the samples also revealed very high average SCC value of these samples (4.05 lakh cells/ml) which were clearly indicative of infection [8], which are also in agreement with the reports of Sharma et al. [8] and Smith [9]. Again, Langer et al. [18] and Shitandi et al. [19] also reported average SCC values of 3-9 lakh cells/ml of milk in their study of bovine subclinical mastitis from Bikaner and Kenya.
A total of 23 (6.5%) samples were found to be typically Pseudomonas sp. as per Carter and Wise [11], Quinn et al. [10] and Samanta [12]. The slightly variable samples were also previously reported by Freitas and Barth [20] in their studies.
Only 19 (5.4%) isolates were confirmed to be P. aeruginosa showing characteristic pyoverdin associated blue-green fluorescence which was also reported by Quinn et al. [10], Samanta [12], and Narayanan [6]. Association of P. aeruginosa in bovine subclinical mastitis cases were also reported earlier in Gujarat [4] and Jammu and Kashimr [5] at 3.6% and 9.4% respectively, which are more or less matching with this study (5.4%). Other workers such as Heleili et al. [21] (3.0%) and Vishwakarma [22] (6.9%) also reported such prevalence of Pseudomonas spp. in subclinical mastitis cases of bovines and buffaloes.
The presence of toxic gene like toxA (63.2%) was more than that of exoS (36.8%) among all 19 positive P. aeruginosa isolates were also reported by Lanotte et al. [13] who detected toxA (approximately 100%) and exoS (84.5%) genes in 162 P. aeruginosa isolates. The occurrence of “exotoxin A” (90-100%) and “exoenzyme S” (50-84%) in P. aeruginosa strains was also reported by Badr et al. [23], Nikbin et al., [24] and Sharma et al. [25].
Conclusion
It can be concluded that approximately 5.4% of the subclinical mastitis cases of bovines in different districts of mainly South Bengal, might be caused by pathogenic P. aeruginosa strains which possess pathogenic genes, viz., toxA (63.2%) and exoS (36.8%). As this pathogen can act as opportunistic one for the human beings also, so proper care should be taken to check this infection.
Authors’ Contributions
SB, SM, KB, SD, and DPI prepare the study design and carried out the experiment. SB, TKS, and SD conducted the molecular part of the research. KB, IS, and SNJ analyzed the data, drafted and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors cordially thank the Vice-Chancellor, the DREF, the Dean, F/VAS, West Bengal University of Animal and Fishery Sciences and the Department of Animal Resources Development and related Government officials of West Bengal who approved and funded (ICAR Development Fund 2013-14) this research work and the farm owner, farmers, local veterinary doctors, and Pranibandhu (Animal Attendants) of different Districts of West Bengal who helped without hesitation during this research work.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.N.D.D.B. Statistics. Milk Production in India. IST; 2013. 2013. [Accessed on 22-10-2013]. Available from: http://www.nddb.coop/English/Statistics/Pages/Milk-Production.aspx .
- 2.Kumar M, Goel P, Sharma A, Kumar A. In: Compendium of 27th. Tamil Nadu, India: ISVM International Summit and Convention at Chennai; 2009. pp. 4–6. [Google Scholar]
- 3.Dua K. Incidence, etiology and estimated economic losses due to mastitis in Punjab and in India - An update. Indian Dairyman. 2001;53(10):41–48. [Google Scholar]
- 4.Patel J.V, Bhingaradia B.V, Patel B.B, Patel S.B, Patel P.B, Vahora S.P. Study on prevalence of mastitis and antibiotic sensitivity of bacterial isolates recovered from crossbred cows of Anand district of Gujarat. Indian J. Dairy Sci. 2012;65(6):467–471. [Google Scholar]
- 5.Singh R, Sharma N, Soodan J.S, Sudhan N.A. Etiology and sensitivity of bacterial isolates from sub-clinical mastitis in cattle from Jammu region. SKUAST J. Res. 2005;4(2):223–224. [Google Scholar]
- 6.Narayanan S. Pseudomonas. In: McVey D.S, Kennedy M, Chengappa M.M, editors. Veterinary Microbiology. 3rd ed. New York: John Wiley & Sons, Inc; 2013. pp. 148–150. [Google Scholar]
- 7.Chakrabarti A. A Textbook of Preventive Veterinary Medicine. 4th ed. New Delhi: Kalyani Publishers; 2007. pp. 477–480. [Google Scholar]
- 8.Sharma N, Singh N.K, Bhadwal M.S. Relationship of somatic cell count and mastitis: An overview. Asian Australas. J. Anim. Sci. 2011;24:429–438. [Google Scholar]
- 9.Smith K.L. Standards for Somatic Cells in Milk: Physiological and Regulatory. Mastitis Newsletter, Newsletters of the IDF No. 144. 1996:7–9. [Google Scholar]
- 10.Quinn P.J, Markey B.K, Leonard F.C, Fitz Patrick E.S, Fanning S, Hartigan P.J. Veterinary Microbiology and Microbial Diseases. 2nd ed. Ames, IA: Blackwell Publishing Ltd; 2011. pp. 287–290. [Google Scholar]
- 11.Carter G.R, Wise D.J. Essentials of Veterinary Bacteriology and Mycology. 6th ed. Iowa: The Iowa State Press; 2004. pp. 125–126. [Google Scholar]
- 12.Samanta I. In: Veterinary Bacteriology. 1st ed. Ch. 10. New Delhi: New India Publishing Agency; 2013. Pseudomonas and Burkholderia; pp. 209–223. [Google Scholar]
- 13.Lanotte P, Watt S, Mereghetti L, Dartiguelongue N, Rastegar-Lari A, Goudeau A, Quentin R. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J. Med. Microbiol. 2004;53:73–81. doi: 10.1099/jmm.0.05324-0. [DOI] [PubMed] [Google Scholar]
- 14.Brenner D.J, McWhorter A.C, Knutson J.K, Steigerwalt A.G. Escherichia vulneris: A new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harini H, Sumathi B.R. Screening of bovine milk samples for sub-clinical mastitis and antibiogram of bacterial isolates. Vet. World. 2011;4(8):358–359. [Google Scholar]
- 16.Marschke R.J, Kitchen B.J. Detection of bovine mastitis by bromothymol blue pH indicator test. J. Dairy Sci. 1985;68:1263–1269. doi: 10.3168/jds.S0022-0302(85)80955-3. [DOI] [PubMed] [Google Scholar]
- 17.Singh R, Bansal B.K, Uppal S.K, Malik D.S. Diagnosis of sub-clinical mastitis: A comparative study of different tests. Indian J. Anim. Res. 2000;34(2):120–123. [Google Scholar]
- 18.Langer A, Sharma S, Sharma N.K, Nauriyal D.S. Comparative efficacy of different mastitis markers for diagnosis of sub-clinical mastitis in cows. Int. J. Appl. Sci. Biotechnol. 2014;2(2):121–125. [Google Scholar]
- 19.Shitandi A, Ogollah H, Nanua J.N. Effect of sub-clinical mastitis on milk composition in the Kenyan smallholder dairy herds. Afr. Crop Sci. Conf. Proc. 2005;7:545–550. [Google Scholar]
- 20.Freitas A.L, Barth A.L. Typing of Pseudomonas aeruginosa from hospitalized patients: A comparison of susceptibility and biochemical profiles with genotype. Braz. J. Med. Biol. Res. 2004;37:77–82. doi: 10.1590/s0100-879x2004000100011. [DOI] [PubMed] [Google Scholar]
- 21.Heleili N, Ayachi A, Melizi M, Kassah A.L, Mamache B. Prevalence of sub-clinical bovine mastitis and the in vitro sensitivity of bacterial isolates in Batna governorate, East of Algeria. J. Anim. Sci. Adv. 2012;2(6):576–582. [Google Scholar]
- 22.Viswakarma P. Studies on Prevalence, Diagnosis, Therapy and Control of Mastitis in Buffaloes. M. V. Sc. Thesis. Raipur, Chhattisgarh: Indira Gandhi Agricultural University; 2008. [Google Scholar]
- 23.Badr H.A.R, El-Nagdy M, El-Sabagh A, El-Din A.B. Pseudomonas aeruginosa exotoxin aas a virulence factor in burn wound infections. Egypt. J. Med. Microbiol. 2008;17(1):125–132. [Google Scholar]
- 24.Nikbin V.S, Aslani M.M, Sharafi Z, Hashemipour M, Shahcheraghi F, Ebrahimipour G.H. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iran. J. Microbiol. 2012;4(3):118–123. [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma S, Kaur R, Yadav V, Harjai K, Joshi K. Contribution of exotoxin A of Pseudomonas aeruginosa in acute & chronic renal infection. Jpn. J. Infect. Dis. 2004;57:119–120. [PubMed] [Google Scholar]


