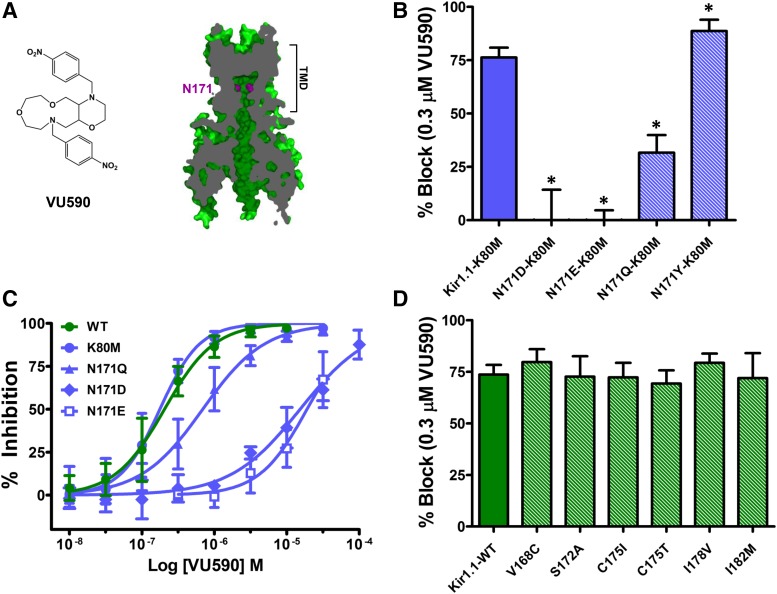
Fig. 1.
N171 is required for VU590 block of Kir1.1. (A) (Left) Chemical structure of VU590. (Right) Section through a Kir1.1 homology model at the level of the pore showing the location of N171 (magenta), which is required for VU590 sensitivity. (B) Percent inhibition by 0.3 μM VU590 of Kir1.1-K80M (solid blue bar) and N171 mutants (striped bars). (C) VU590 CRCs for Kir1.1-WT (wild-type), K80M, N171Q, N171D and N171E mutants. (D) Percent inhibition of Kir1.1-WT (solid green bar) and respective mutants (striped bars) by 0.3 μM VU590. Data were analyzed using one-way ANOVA with Bonferroni’s multiple comparison test and plotted as mean ± S.D. percent inhibition (n ≥ 4); *P < 0.05.