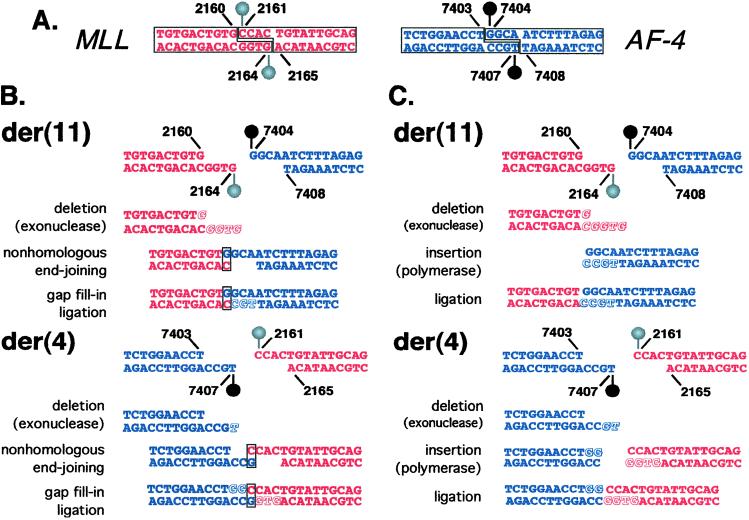
Figure 3.
(A) DNA topoisomerase II cleavage sites identified in vitro. The DNA topoisomerase II homodimer creates four-base staggered nicks in duplex DNA. Cleavage assays indicated that MLL position 2,160 is at the 5′ side (−1 position) of a cleavage site on the sense strand. The corresponding cleavage site on the antisense strand staggered by four bases from MLL position 2,160 is position 2,164 (Left, red). Cleavage assays indicated that AF-4 position 7,403 is at the 5′ side (−1 position) of a cleavage site on the sense strand. The corresponding cleavage site on the antisense strand staggered by four bases from position AF-4 position 7,403 is position 7,407 (Right, blue). (B and C) Models for processing of four-base 5′ overhangs of DNA topoisomerase II cleavage sites in MLL and AF-4 to generate observed der(11) and der(4) genomic breakpoint junctions. (B) To form the der(11), one possibility on the MLL side is that the entire four-base 5′ antisense-strand overhang (positions 2,161–2,164) and G at position 2,160 on the sense strand are removed by exonucleolytic nibbling (Red, italic, shadowed). On the AF-4 side, the entire four-base 5′ sense-strand overhang is preserved. The exonucleolytic nibbling of the MLL overhang would create a single-base microhomology, which facilitates base-pairing between the C at MLL antisense position 2,160 and the G at position 7,404 in the AF-4 sense-strand overhang (box). Template-directed polymerization of the antisense strand fills in the gap at AF-4 positions 7,107–7,105 (Blue, shadowed, nonitalic). The der(11) forms by fusion of position 2,159 on the MLL sense strand to position 7,404 on the AF-4 sense strand. To form the der(4) junction, exonucleolytic nibbling on the AF-4 side removes position 7,407 from the four-base 5′ antisense-strand overhang (Blue, italic, shadowed), whereas the entire four-base MLL 5′ sense-strand overhang remains intact. The exonucleolytic nibbling of the AF-4 overhang would create a single-base microhomology, and there is base-pairing between the G at AF-4 antisense position 7,406 and the C at position 2,161 of the MLL sense-strand overhang (box). Template-directed polymerization fills in gaps at AF-4 sense positions 7,404–7,405 (Blue, shadowed, nonitalic) and MLL antisense positions 2,164–2,162 (Red, shadowed, nonitalic). The der(4) forms by fusion of position 7,405 on the AF-4 sense strand to position 2,161 on the MLL sense strand. There are three G residues in the normal MLL and AF-4 sense sequences that form the breakpoint junctions at MLL position 2,160 and at AF-4 positions 7,404 and 7,405. During the complex processing, the G at MLL position 2,160 is removed; both G residues at AF-4 positions 7,404 and 7,505 are preserved at the der(11) junction; and both G residues at AF-4 positions 7,404 and 7,505 are added by gap fill-in at the der(4) junction (compare box, Fig. 1), resulting in an overall gain of a single G relative to the normal MLL and AF-4 sense sequences. (C) Compared with B, there is more exonucleolytic nibbling in this model. The entire four-base 5′ antisense-strand overhang, the G at position 2,160 on the sense strand, and the complementary C are deleted from the MLL end (Red, italic, shadowed) that forms the der(11). Positions 7,406–7,407 of the 5′ antisense-strand overhang are deleted from the AF-4 end (Blue, italic, shadowed) forming the der(4). After template-directed polymerization (Blue, Red, shadowed, nonitalic), ligations at both junctions are blunt-ended. The der(11) forms by fusion of MLL position 2,159 to AF-4 position 7,404. The der(4) forms by fusion of AF-4 position 7,405 to MLL position 2,161. This model is consistent with the deletion of a single G from MLL to form the der(11) and the gain of two templated Gs in AF-4 to form the der(4), resulting in an overall gain of a single G relative to the normal MLL and AF-4 sense sequences. Because of the required processing, simple reciprocal exchange of DNA topoisomerase II subunits alone could not produce the observed der(11) and der(4) breakpoint-junction sequences.