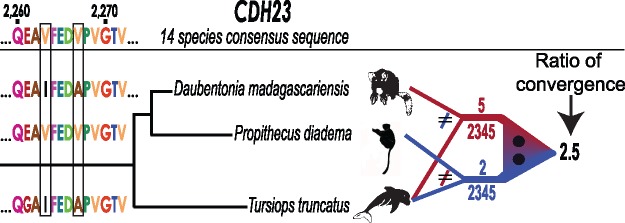
Fig. 2.—
Illustration of the pairwise convergent evolution analysis used in this study. The example shown is for a section of the CDH23 gene, here comparing two lemurs, Daubentonia and Propithecus diadema, with the echolocating dolphin Tursiops truncatus. For each lemur, we computed the number of positions out of the total aligned positions (2,345 for this gene) for which that species shared an amino acid with T. truncatus, but for which that amino acid was different than that of the other lemur species and the multi-species consensus sequence determined from the multiple-species alignment. A section of CDH23 is shown that contains two Daubentonia—T. truncatus convergent amino acids. For the whole gene, the ratio of convergent amino acids for Daubentonia and T. truncatus (5) and P. diadema and T. truncatus (2) = 2.5, providing a magnitude and directionality of relative convergence between aye-aye and dolphin, with a phylogenic correction based on the sister lemur species. These ratio values are depicted in figure 3 for all tested comparisons, summed across the seven genes analyzed in this study. The difference in the number of convergent amino acids is evaluated with a Fisher’s exact test computed based on the 2×2 contingency table, not the convergence ratio; for the illustrated CDH23 comparison, P = 0.4528, prior to Bonferroni multiple test correction.