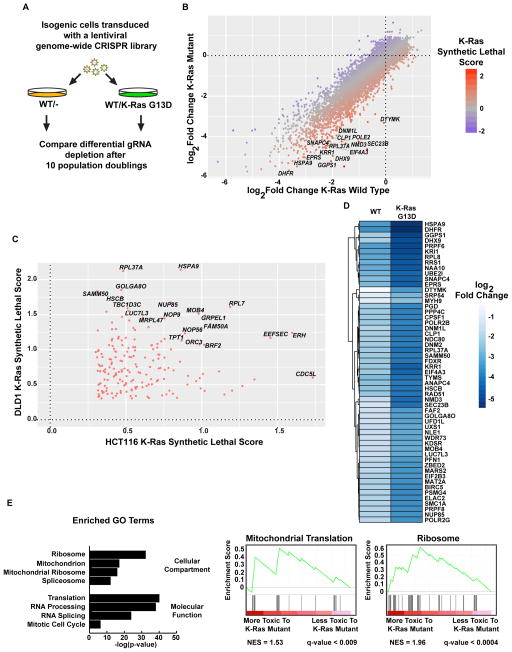
Figure 1. Genome-wide CRISPR screens in isogenic colorectal tumor cells identifies synthetic lethal interactions with mutant K-Ras.
A. Schematic of the CRISPR screens performed. Abundance of gRNAs was compared between the initial (PD0) and final (PD10) time points. Genes whose gRNAs are selectively depleted in the K-Ras mutant cells are putative synthetic lethal hits. B. Fold changes of genes in the DLD1 screen. The average log2 fold change for each gene was calculated and plotted for both WT/- and WT/K-Ras mutant DLD1 cells. Labeled genes are a selection of potential synthetic lethal partners of mutant K-Ras. C. Synthetic lethal genes that scored in both the HCT116 and DLD1 screens. D. Average fold changes for the top 55 K-Ras synthetic lethal genes identified in the DLD1 screen. E. Gene ontology terms enriched in putative K-Ras synthetic lethal genes. For screen sequencing data and analyses, see Supplemental Tables S1–4.