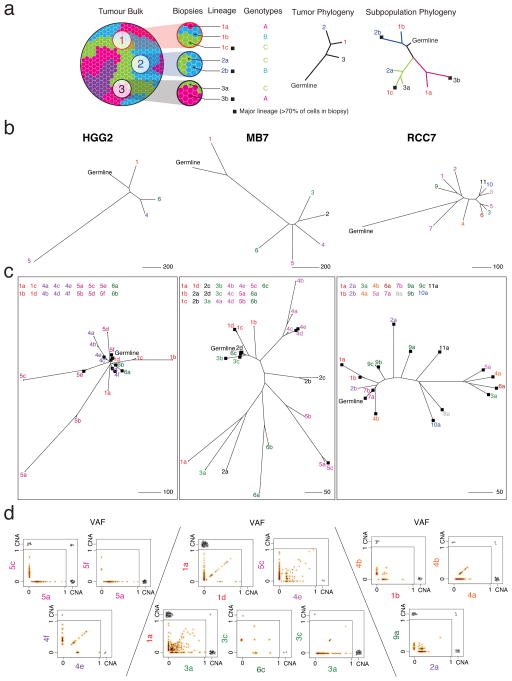
Figure 3. Spatial intermixing of clonal lineages.
(a) Example cartoon of a tumor with four clonal lineages that are spatially dispersed (blue, green, pink, purple) demonstrates how data from three biopsies are used to build a typical biopsy-level phylogenetic tree as well as a subpopulation-level tree reflecting inter-mixing of the three detected genetic lineages. Branch tips are colored according to biopsy number and labeled according to biopsy number (1,2,3…) and clonal lineage (a,b,c…). Branch colors correspond to the cellular genotype; black squares indicate major cellular lineages (>70% of tumor cells in the biopsy, scaled by the largest detectable population). Note that the number of biopsies may not be sufficient to ‘discover’ all distinct clonal lineages (e.g. purple clone). (b) Biopsy-level trees of three representative tumors; MB7, HGG2, and RCC7. (c) Subpopulation-level trees reveal that some cellular lineages have high similarity to lineages in other biopsies, suggesting spatial intermixing (e.g. MB7 biopsy 1,2,3; RCC7 biopsy 4). Conversely, some biopsies harbor >1 distinct lineage (e.g. HGG2 biopsy 5). (d) Variant allele frequency (VAF) of mutations are shown along with copy number aberrations exclusive to or shared by pairs of biopsies or subpopulations. VAF scatter plots have a smoothed color density; black dots represent individual mutations. CNA events (black triangles) are displayed (with some jitter) if present in either compartment, or shared.