Abstract
Kallikrein-related peptidases (KLKs) are crucial for epidermal barrier function and are involved in the proteolytic regulation of the desquamation process. Elevated KLK levels were reported in atopic dermatitis. In skin, the proteolytic activity of KLKs is regulated by specific inhibitors of the serine protease inhibitor of Kazal-type (SPINK) family. SPINK6 was shown to be expressed in human stratum corneum and is able to inhibit several KLKs such as KLK4, -5, -12, -13 and -14. In order to understand the structural traits of the specific inhibition we solved the structure of SPINK6 in solution by NMR-spectroscopy and studied its interaction with KLKs. Thereby, beside the conserved binding mode, we identified an alternate binding mode which has so far not been observed for SPINK inhibitors.
Keywords: SPINK6, kallikrein-related peptidase 4, KLK, structure, model protease – inhibitor complex, nuclear magnetic resonance
1. Introduction
Proteases are responsible for the maintenance of the epidermal barrier by proteolytic cleavage of filaggrin and other desmosomal proteins. Among these proteases, the kallikrein-related peptidases (KLKs) are important in the proteolytic regulation of the desquamation process. Members of this family are involved in many of these processes and their insufficient regulation can lead to severe diseases [1].
Kallikrein-related proteases (KLKs) belong to the large class of tryptic and chymotryptic serine proteases. In humans, the KLK family encompasses 15 genes encoded within the single chromosomal locus 19q13.3–13.4, whereas in mice and rat the KLK family consists of 24 and 20 genes, respectively [2]. At least eight KLKs are expressed in human skin [3]. KLK4 has been detected at the mRNA level in human skin, and at the protein level in healthy adult and fetal skin samples [4–6]. In skin, the enzymatic activity of KLKs is mainly regulated by specific inhibitors of the serine protease inhibitor of Kazal-type (SPINK) family [7]. Spink6, with a single Kazal-type domain, is well conserved among homologs from frogs to humans, suggesting an important role in cellular function [8]. KLK4 was described in human skin, but its functional role in skin biology is poorly understood [5,9]. In the stratum corneum of the human skin SPINK6 is cross-linked inter alia to fibronectin by transglutaminases [10,11]. SPINK6 inhibits members of the KLK family with different efficacies. These can be grouped in high (KLK5, 12, 13 and 14), medium (KLK4) and low affinity (KLK6 and 7) inhibition [12]. The medium inhibition of KLK4 activity by SPINK6 is reflected in a Ki value of 27 nM [12]. In order to elucidate the molecular basis of the KLK4 inhibition by SPINK6 we solved the structure of SPINK6 by NMR-spectroscopy and generated a model of its complex with KLK4 in order to identify the interaction epitope.
2. Materials and methods
2.1. Protein expression and purification
SPINK6 was expressed and purified as described before [10].
2.2. NMR spectroscopy
For structure determination by NMR-spectroscopy SPINK6 was labeled with 15N- and 13C-isotopes by expression in minimal media supplemented with 2 g/l 15C-glucose and 1 g/l 15N-ammonium sulfate, and purified as described for the non-labeled material. All spectra were processed with NMRPipe [13] and analyzed with NMRViewJ [14]. NMR measurements were performed on a Bruker Avance 600-MHz spectrometer equipped with a z-gradient triple-resonance cryoprobe. Sequence-specific backbone resonance assignments of SPINK6 were established using the following spectra: 1H–15N-heteronuclear-HSQC, three-dimensional, (H)C(CO)NH, H(CCO)NH, HN(CO)CACB, HNCACB, 15N-edited TOCSY, and 13C-edited HCCH-TOCSY. For verification of flexible regions (1H)–15N heteronuclear nuclear Overhauser effect (NOE) spectra were measured additionally. Two spectra with on and off resonance 1H saturation were recorded in an interleaved manner, and separated before processing with the Bruker AU-program split. The combined recycle and saturation delay was set to 3.2 s. Only well resolved peaks were included in the intensity (peak height) measurements and NOE ratio determination. Distance restraints were obtained from three-dimensional 15N-edited and 13C-edited NOE spectra (mixing times, 150 ms).
The spectra were acquired at 303 K and referenced to the water resonance at 4.79 ppm. Chemical shifts and constraints were deposited in the BMRB, accession number: 25691.
2.3. Structure calculation
Structure calculations were performed using the program CYANA [15]. The structure calculation was based on 450 interproton distances derived from 15N and 13C-edited three-dimensional NOESY experiments. Distances restraints were calibrated using an r6 function. Three disulfide bonds were defined as 9 distance restraints ranges as follows: 2.0 ≤ d(Siγ, Sjγ) ≤ 2.1 Å; 3.0 ≤ d(Ciβ, Sjγ) ≤ 3.1 Å; 3.0 ≤ d(Siγ, Cjβ) = 3.1 Å. TALOS+ [16,17] was used for deriving backbone torsion angles from the NMR chemical shifts of SPINK6 and included in the structure calculations. 100 structures were calculated and subsequently refined in explicit solvent with the CNS program using the RECOORD protocol and parameters [18]. The 20 energetically best structures with an NOE violation energy below a cutoff were selected as the final refined structural ensemble and were deposited (protein data bank accession code: 2n52) (Table 1). All molecular graphical representations were generated using the program RIBBONS [19].
Table 1.
Structural statistics for the 20 best structures of SPINK6. No distance constraint was violated by more than 0.5 Å in any structure. Rmsd = root mean square deviation. *, rmsd was calculated for regions containing helix and strands.
| Distance restraints | Number |
|---|---|
| Intraresidual (|i-j| = 0) | 173 |
| Sequential (|i-j| = 1) | 161 |
| Medium range (2 ≤ |i-j| ≤ 4) | 57 |
| Long range (|i-j| ≥ 5) | 59 |
| Three disulphide bonds included | 18 |
| Total | 450 |
| Pairwise rmsd* | |
| Average rmsd backbone all | 0.59 ± 0.12 |
| Heavy atoms rmsd: | 1.25 ± 0.19 |
| Ramachandran plot (for average structure out of 20 structures) | |
| Most favored regions (%) | 77 |
| Additional allowed regions (%) | 23 |
| Generously allowed regions (%) | 0.5 |
| Disallowed regions (%) | 0.1 |
2.4. Model building
The region of SPINK6 that is in contact with the protease, KLK4, is very flexible in solution, thereby imposing challenges upon the modeling of the SPINK6-KLK4 complex. To tackle this challenge, we exploited a template-restrained docking protocol, which has been shown to systematically improve superimposed models (Xue, Rodrigues, Dobbs, Honavar, & Bonvin, submitted). This protocol is based on the flexible protein–protein docking method, HADDOCK [20,21] which can incorporate user-provided interface information, either experimental or predicted. Using interface restraints, HADDOCK is able to perform a more detailed flexible refinement of the interface and generate near-native conformations even for cases with medium to large conformational changes [22]. To identify available templates for SPINK6-KLK4 in the PDB (Protein Data Bank, www.wwpdb.org) [23], we used PS-HomPPI [24]. Given the sequences of two interacting query proteins, the newest version of PS-HomPPI -- v1.3 searches homologous interacting proteins in the PDB, classifies the templates into Safe Zone (high level of interface conservation), Twilight Zone (medium level) or Dark Zone (low level), clusters the templates based on the structural similarity and returns the inferred clustered interfaces from the templates in the best available zone. For SPINK6-KLK4, PS-HomPPI v1.3 identified 8 templates (Table 2), all of them in the Twilight Zone.
Table 2.
Eight templates identified by PS-HomPPI v1.3.
| PDB ID | CHAIN ID | IDENTITY with SPINK6 | IDENTITY with KLK4 |
|---|---|---|---|
| 1ppf | I:E | 55.56 | 31.58 |
| 1hja | I:B | 56.60 | 34.85 |
| 1z7k | B:A | 33.33 | 39.11 |
| 1cho | I:F | 55.56 | 34.85 |
| 1cho | I:G | 55.56 | 36.46 |
| 1ldt | L:T | 41.46 | 39.11 |
| 1an1 | I:E | 41.46 | 39.11 |
| 1tgs | I:Z | 36.59 | 40.44 |
Reliable templates for SPINK6-KLK4 could be identified (with reasonably high sequence identity ranging from 32% to 56%). Superimposed models for SPINK6-KLK4 using the top template identified by PS-HomPPI v1.3: an x-ray crystallography solved complex between a human leukocyte elastase and OMTKY3 (the third domain of turkey ovomucoid inhibitor), PDB ID: 1ppf. These were then refined skipping the initial rigid-body docking stage (it0) and performing only the semi-flexible refinement (it1 and water) of HADDOCK v2.2 with default parameters. The following unambiguous restraints: were used in the refinement: a) CA–CA (C alpha – C alpha) distances calculated by PS-HomPPI v1.3 from the 8 templates in Table 2, b) H-bonds from the template 1ppf, c) dihedral angles extracted from 1ppf to restrain the conformation of the inhibitory loop to the bound form, d) the original NMR NOE restraints, and e) distance restraints extracted from the KLK4 structure to keep the catalytic triad (Asp-His-Ser) in KLK4 in place, whose position is known to be highly conserved. The N-terminus tail (residue 1–22) of SPINK6 was treated as fully flexible during the it1 and water refinement stages. From the 200 calculated SPINK6-KLK4 models we chose top 10 based on HADDOCK scores. These models were further subjected to a refinement round in explicit solvent, without any restraints (using the refinement interface of the HADDOCK server) to remove any strain introduced by the restraints.
3. Results
3.1. Structure of SPINK6
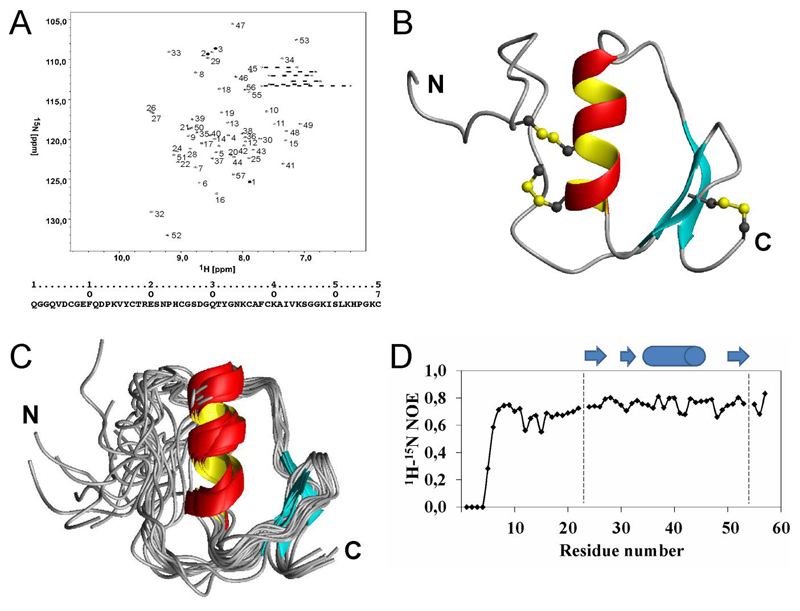
The 1H–15N-HSQC spectrum and the sequential assignment of SPINK6 is depicted in Fig. 1A showing good dispersion of the resonances. All residues could be unambiguously assigned. 450 distance constraints were derived from the 15N- and 13C-edited NOESY spectra and used to calculate the three-dimensional structure. Fig. 1B shows the representative overall structure of SPINK6. From 100 calculated structures, the 20 energetically best structures were selected for the ensemble which is shown in Fig. 1C. The structural statistics of this ensemble are shown in Table 1. The overall structure of SPINK6 comprises two adjacent β-strands, followed by an α-helix and a C-terminal β-strand. All secondary elements are well defined as it can be deduced from the ensemble. Since the N-terminus is less defined and exhibited a wide range of different conformations we assessed the backbone flexibility by 1H–15N heteronuclear NOE experiments. Fig. 1D shows the analysis of these data. Whereas for residues 7 to 57 the values around 0.8 are indicative of a rigid structure, lower NOE ratios for the N-terminal part (residue 1 to 6) confirm the high flexibility and lack of structure in accordance with the results of the structure calculations.
Figure 1.
(A) 1H–15N HSQC-spectrum of SPINK6. Resonances are numbered according to the sequence numbers below. Side chain H–N resonances are connected by dotted lines. (B) Ribbon representation of the SPINK6 structure. Side chains of the disulfide-bridges are depicted in yellow and gray and the N-terminus (N) and C-terminus (C) are labeled. (C) Overlay of the final refined 20 structures ensemble. For superposition the secondary structural elements were used and the N-terminus (N) and C-terminus (C) are labeled. (D) Flexibility of the protein backbone measured by heteronuclear NOEs. Positions of helix (cylinder) and beta-strands (arrows), respectively, are depicted on top according to the involved residue numbers.
3.2. Structural comparison of SPINK6 with SPINK2 and SPINK7
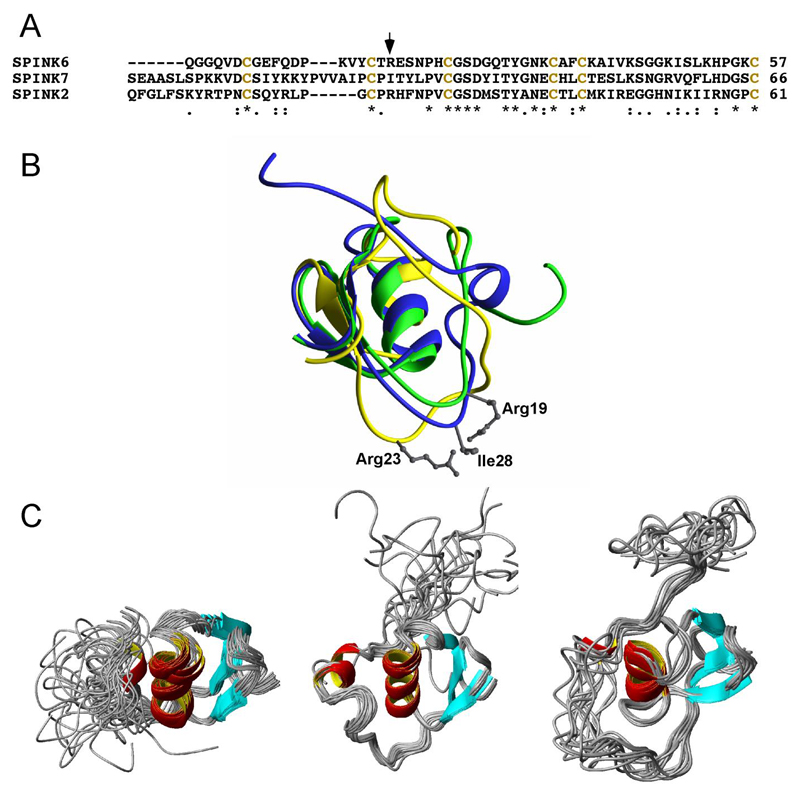
The structures of two other members of the SPINK family have been previously solved – i.e. SPINK2 [25] and SPINK7 [26]. Fig. 2A shows the sequence alignment of the three molecules. The cysteine pattern is conserved in all three molecules. The central residue suggested to be involved in binding to the so-called S1-pocket of the active site of their target proteases is an arginine in case of SPINK6 and 2, and an isoleucine in case of SPINK7. Arginine residues at this position are typical for trypsin-like proteases, whereas bulky hydrophobic residues, like isoleucine, are typical for chymotrypsin-like proteases. Fig. 2B shows the superposition of the three representative SPINK structures. Whereas the central α-helix and the β-sheet are structurally conserved, the N-termini differ remarkably. As a consequence, the residues of the SPINK molecules with the S1-pocket of the active site of the proteases differ significantly in their orientation. Fig. 2C shows the side-by-side comparison of the three ensembles of the SPINK structures. Again, the central α-helix and the β-sheet are very well defined, whereas the N-termini of all three molecules are highly flexible und unstructured.
Figure 2.
Comparison of the structures of SPINK2, 6, and 7. (A) Sequence alignment of SPINK2, 6, and 7. The position of residues known to bind to the active site of the proteases is marked by an arrow. (B) Superposition of the averaged structures of SPINK6 (green), SPINK2 (yellow, PDB ID: 2jxd) and SPINK7 (blue, PDB ID: 2leo). For superposition the secondary structure elements were used. Side chains of the residues known to bind to the active site of the S1-pocket of the proteases are depicted and labeled. (C) Structural ensembles of SPINK6, SPINK7 and SPINK2 (from left to right). In all three molecules, the N-terminal region, which contains the amino-acid residue side chain involved in the inhibition of KLKs, exhibits a high flexibility. Molecules are shown in the orientation with respect to the central helix.
3.3. The SPINK6 – KLK4 model
Using the information derived from the identified templates (see Material and Methods) as restraints to define the backbone conformation of the interaction loop and to maintain the conserved arrangement of the catalytic triad in SPINK6, we were able to generate models of the complex of SPINK6 and KLK4, which show an improved conformation of the interaction loop of SPINK6 (Fig. 3).
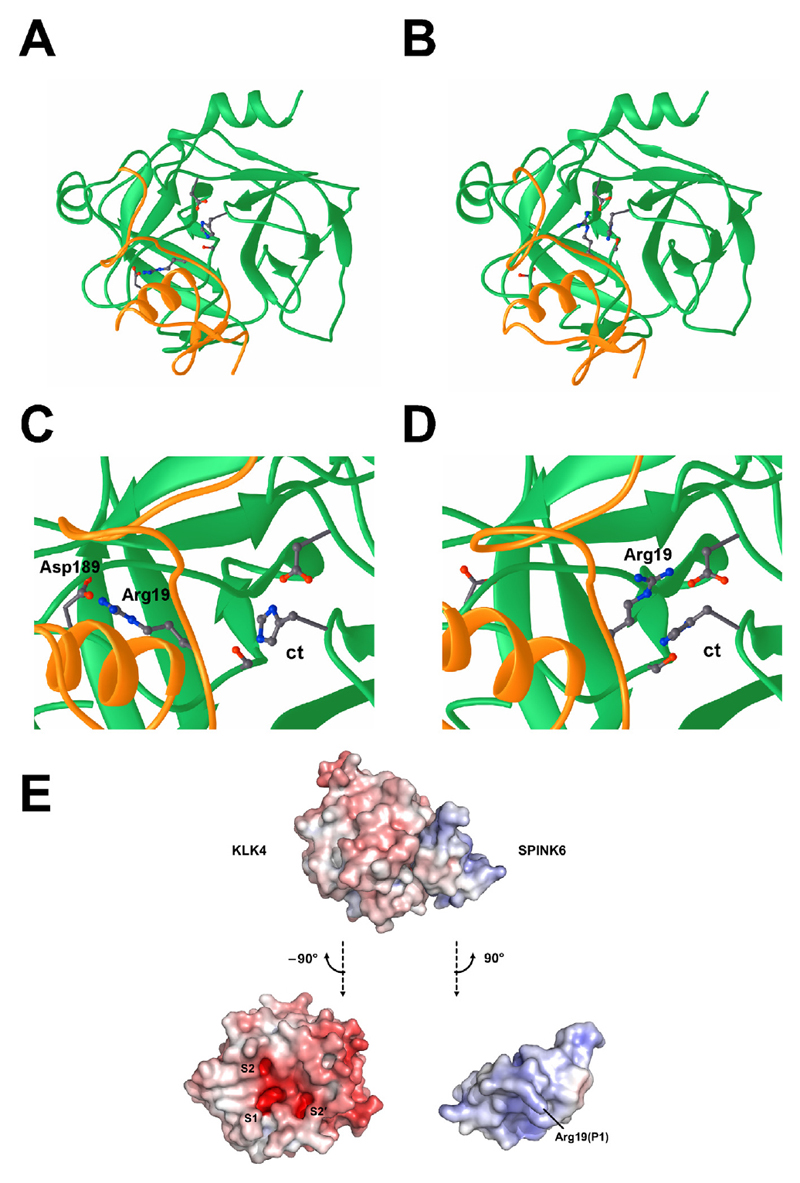
Figure 3.
The complex of SPINK6 (yellow) and KLK4 (green). (A and B) Overview of the two conformations of the KLK4-SPINK6 complex. The amino-acid residues constituting the active site of the protease (catalytic triade, ct, Asp102/His57/Ser195), the canonical binding residue (Asp189) and Arg19 of SPINK6 are depicted. (C and D) Close up of the two alternative binding modes of Arg19 of SPINK6 to the active site of KLK4. (E) The KLK4-SPINK6 complex in the canonical binding mode, shown sideways with respect to the standard orientation of the protease. The electrostatic potential is contoured from −10 kBT/e (red) to +10 kBT/e (blue). In the lower panel, both KLK4 and SPINK6 have been turned by 90°, but in opposite directions. The KLK4 active site exhibits highly negative potentials in the S1, S2, and S2′ specificity pockets. Additionally, an extended negatively charged patch on the right might serve as exosite for substrate binding. The SPINK6 reactive centre loop and surrounding region is both complementary to KLK4 with respect to shape and its positively charged residues, such as Arg19, which is flexible enough to switch between binding of the S1 and S2 pockets.
The interaction loops of refined SPINK6 models (residues 17–22) are now forming conformations that are similar to the templates; furthermore, the previously disordered N-termini (residues 1–16) of unbound SPINK6 NMR models, which originally pointed in various directions and initially clashed with KLK4 after superimposition, do not clash any more with KLK4. The resulting models still satisfy the original NOE restraints.
While the backbone interaction conformations of the top 10 models are almost identical, the side chain of the arginine (Arg19) known to bind the active site of KLK4 presents two reasonable but distinct binding conformations1 (Fig. 3): 1) conserved binding mode: binding the primary pocket of KLK4 as in the templates (Fig. 3C); 2) an alternate binding mode: binding the secondary S2 pocket (Fig. 3D). Models with either binding conformation have reasonable HADDOCK energy score (Table 3).
Table 3.
HADDOCK statistics for SPINK6-KLK4 models grouped with two distinct conformations of the Arg19 side chain. Evdw is Van der Waals energy, Eelec is electrostatic energy, and BSA is Buried Surface Area.
| Arg_19 (conserved mode) | Arg_19 (alternate mode) | |
|---|---|---|
| Evdw [kJ mol-1] | -325.1 ± 13.4 | -282.0 ± 14.2 |
| Eelec [kJ mol-1] | -725.9 ± 77.0 | -979.1 ± 231.4 |
| BSA [Å2] | 1948 ± 53 | 1861 ± 57 |
| HADDOCK score [a.u.] | -113.2 ± 3.7 | -114.9 ± 6.2 |
4. Discussion
Proteolytic processes play an important role in many physiological and pathophysiological situations. The members of the large superfamily of serine proteases are involved in blood coagulation, activation of the complement system, but also cancer progression. Several kallikrein-related peptidases has been described to be involved in the proteolytic regulation of the desquamation process of the skin. In addition to their physiological roles, the dysregulation of KLK expression, activity or localization, has been described to be associated with pathophysiological disorders, such as skin disorders, respiratory diseases, perturbation of tooth enamel development, neurological disorders and carcinogenesis [27].
The proteolytic activities of these proteases are regulated by natural inhibitors such as metal ions, serpins, macroglobulins, and Kazal-type inhibitors [4]. Although expression levels of KLK4 in skin are moderate and transient [5], a regulation by SPINK6 seems plausible. However, the major targets of SPINK6 in skin are most likely the two tryptic proteases KLK5 and KLK14, exhibiting high expression levels [10].
Kazal-type inhibitors of trypsin-like serine-proteases share a central basic residue, either a lysine or an arginine residue (corresponding to Arg19 of SPINK6, see Fig. 2A) which targets the aspartic acid (Asp189) at the bottom of the S1 specificity pocket in a substrate-like manner. Usually, the S1 pockets accommodate the basic P1 amino acid of a substrate, which is cleaved by trypsin-like proteases. Interestingly, our results suggest a second alternative mode of interaction. Here, Arg19 of SPINK6 binds to the S2 specificity pocket and interacts with the aspartic acid, Asp102. Although binding to the protease may favor the canonical mode, it is remarkable that the alternative conformation is suitably preformed and complementary to the S2 pocket, which exhibits a high negative electrostatic potential, comparable to the one of the S1 pocket (Fig. 3E). Such an interaction has so far not been observed for SPINK inhibitors. Further structural studies of other SPINK/KLK complexes have to be performed, in order to prove whether such an alternative binding mode really exists and how it might contribute to the different inhibitory effects observed for the different KLKs.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (SFB 877, A6 and ME2037/3-3) and the Excellence Cluster 306 ‘Inflammation at Interfaces’. L.X. acknowledges financial support from the Dutch Foundation for Scientific Research (NWO) (VENI grant #722.014.005). P.G. was supported by the Austrian Science Fund FWF (Project P25003-B21).
Footnotes
Two models were discarded due to an unrealistic conformation of Arg19, which pierced deep into the surface of KLK4.
References
- [1].Fischer J, Meyer-Hoffert U. Regulation of kallikrein-related peptidases in the skin - from physiology to diseases to therapeutic options. Thromb Haemost. 2013;110:442–449. doi: 10.1160/TH12-11-0836. [DOI] [PubMed] [Google Scholar]
- [2].Wines DR, Brady JM, Pritchett DB, Roberts JL, MacDonald RJ. Organization and expression of the rat kallikrein gene family. J Biol Chem. 1989;264:7653–7662. [PubMed] [Google Scholar]
- [3].Hansson L, Backman A, Ny A, Edlund M, Ekholm E, Ekstrand Hammarstrom B, Tornell J, Wallbrandt P, Wennbo H, Egelrud T. Epidermal overexpression of stratum corneum chymotryptic enzyme in mice: a model for chronic itchy dermatitis. J Investig Dermatol. 2002;118:444–449. doi: 10.1046/j.0022-202x.2001.01684.x. [DOI] [PubMed] [Google Scholar]
- [4].Komatsu N, Takata M, Otsuki N, Toyama T, Ohka R, Takehara K, Saijoh K. Expression and localization of tissue kallikrein mRNAs in human epidermis and appendages. J Investig Dermatol. 2003;121:542–549. doi: 10.1046/j.1523-1747.2003.12363.x. [DOI] [PubMed] [Google Scholar]
- [5].Obiezu CV, Shan SJ, Soosaipillai A, Luo LY, Grass L, Sotiropoulou G, Petraki CD, Papanastasiou PA, Levesque MA, Diamandis EP. Human kallikrein 4: quantitative study in tissues and evidence for its secretion into biological fluids. Clin Chem. 2005;51:1432–1442. doi: 10.1373/clinchem.2005.049692. [DOI] [PubMed] [Google Scholar]
- [6].Obiezu CV, Soosaipillai A, Jung K, Stephan C, Scorilas A, Howarth DH, Diamandis EP. Detection of human kallikrein 4 in healthy and cancerous prostatic tissues by immunofluorometry and immunohistochemistry. Clin Chem. 2002;48:1232–1240. [PubMed] [Google Scholar]
- [7].Goettig P, Magdolen V, Brandstetter H. Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs) Biochimie. 2010;92:1546–1567. doi: 10.1016/j.biochi.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fischer J, Wu Z, Kantyka T, Sperrhacke M, Dimitrieva O, Koblyakova Y, Ahrens K, Graumann N, Baurecht H, Reiss K, Schroder JM, et al. Characterization of Spink6 in mouse skin: the conserved inhibitor of kallikrein-related peptidases is reduced by barrier injury. J Investig Dermatol. 2014;134:1305–1312. doi: 10.1038/jid.2013.502. [DOI] [PubMed] [Google Scholar]
- [9].Komatsu N, Saijoh K, Sidiropoulos M, Tsai B, Levesque MA, Elliott MB, Takehara K, Diamandis EP. Quantification of human tissue kallikreins in the stratum corneum: dependence on age and gender. J Investig Dermatol. 2005;125:1182–1189. doi: 10.1111/j.0022-202X.2005.23933.x. [DOI] [PubMed] [Google Scholar]
- [10].Meyer-Hoffert U, Wu Z, Kantyka T, Fischer J, Latendorf T, Hansmann B, Bartels J, He Y, Gläser R, Schröder JM. Isolation of SPINK6 in human skin: selective inhibitor of kallikrein-related peptidases. J Biol Chem. 2010;285:32174–32181. doi: 10.1074/jbc.M109.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fischer J, Koblyakova Y, Latendorf T, Wu Z, Meyer-Hoffert U. Cross-linking of SPINK6 by transglutaminases protects from epidermal proteases. J Investig Dermatol. 2013;133:1170–1177. doi: 10.1038/jid.2012.482. [DOI] [PubMed] [Google Scholar]
- [12].Kantyka T, Latendorf T, Wiedow O, Bartels J, Glaser R, Dubin G, Schröder JM, Potempa J, Meyer-Hoffert U. Elafin is specifically inactivated by RgpB from Porphyromonas gingivalis by distinct proteolytic cleavage. Biol Chem. 2009;390:1313–1320. doi: 10.1515/BC.2009.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- [14].Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- [15].Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- [16].Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- [18].Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Güntert P, Livny M, Markley JL, Nilges M, Ulrich EL, et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- [19].Carson M. Ribbons. Methods Enzym. 1997;277:493–505. [PubMed] [Google Scholar]
- [20].de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- [21].Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- [22].Karaca E, Bonvin AM. A multidomain flexible docking approach to deal with large conformational changes in the modeling of biomolecular complexes. Structure. 2011;19:555–565. doi: 10.1016/j.str.2011.01.014. [DOI] [PubMed] [Google Scholar]
- [23].Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- [24].Xue LC, Dobbs D, Honavar V. HomPPI: a class of sequence homology based protein-protein interface prediction methods. BMC Bioinform. 2011;12:244. doi: 10.1186/1471-2105-12-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen T, Lee TR, Liang WG, Chang WS, Lyu PC. Identification of trypsin-inhibitory site and structure determination of human SPINK2 serine proteinase inhibitor. Proteins. 2009;77:209–219. doi: 10.1002/prot.22432. [DOI] [PubMed] [Google Scholar]
- [26].Feng Y, Geng Y, Zhou T, Wang J. NMR structure note: human esophageal cancer-related gene 2. J Biomol NMR. 2012;53:65–70. doi: 10.1007/s10858-012-9622-9. [DOI] [PubMed] [Google Scholar]
- [27].Kalinska M, Meyer-Hoffert U, Kantyka T, Potempa J. Kallikreins - the melting pot of activity and function. Biochimie. 2015 doi: 10.1016/j.biochi.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]