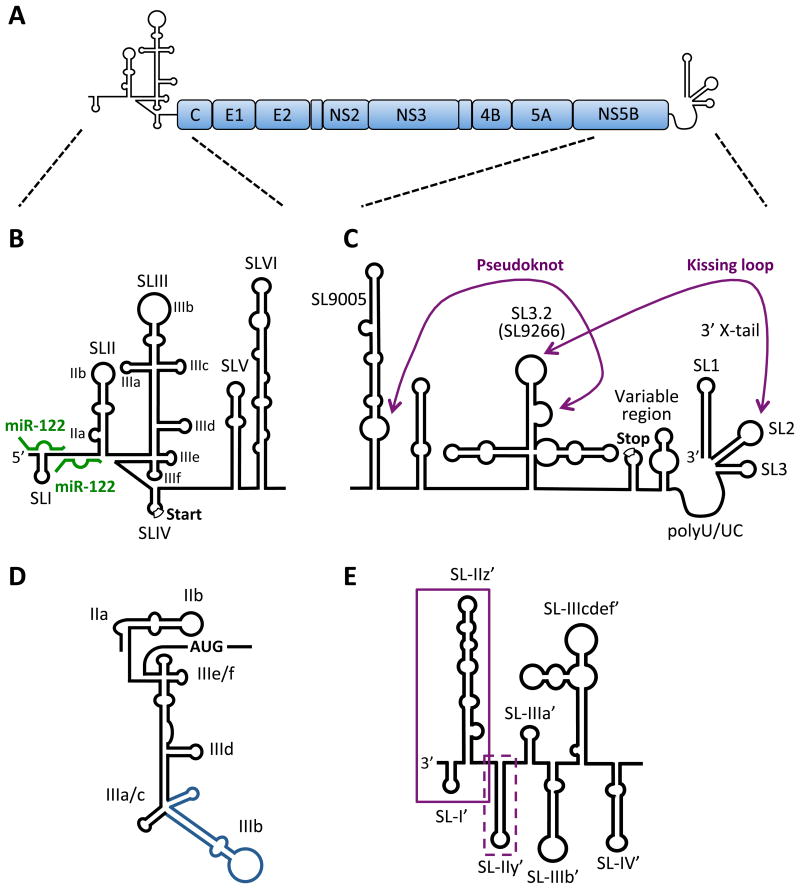
Figure 1. RNA secondary structures in the HCV genome.
A) HCV genome organization. The HCV genome is a positive-sense, single-stranded RNA encoding a single large open reading frame (∼3000 amino acids), flanked by highly structured 5′ and 3′ NCRs. B) Schematic diagram of the 5′ NCR and RNA structures of the core-coding region. The 5′ NCR consists of four stem-loop structures (SLI to SLIV) and the core coding region contains two stem-loop structures (SLV and SLVI). SLII through SLIV comprise the HCV IRES element required for cap-independent translation (start codon is indicated) and SLI through SLII are required for viral RNA replication. The miR-122 sites are indicated (green). C) The 3′ NCR has a tripartite structure containing a variable region (with the polyprotein stop codon), poly-U/UC tract and 3′ × region (containing 3′ SL1, SL2 and SL3). The kissing-loop interaction between 3′ SL2 and SL3.2 (SL9266) of the NS5B-coding region (part of a larger cruciform CRE) as well as a pseudoknot between the 3′ sub-terminal bulge of SL3.2 and residues in the bulge of the extended stem-loop SL9005 (around nt 9110) are indicated. SL nomenclature used is based on the H77 complete genome sequence (Genbank Accession #AF011753). D) The secondary structure of the IRES reflecting its orientation in complex with the 40S subunit and eIF3. SLIIa induces a bent structure in SLII to direct SLIIb to the ribosomal E-site in the head region of the 40S subunits. The SLIIIad junction binds to the surface of the 40S whereas the SLIIIbc motif (blue) associates with eIF3. SLIIIef and the SLIV pseudoknot (not shown) position the AUG codon in the P-site of the 40S subunit. E) The 3′ end of the negative-strand replicative intermediate. Boxed sequences indicate required (solid) and contributing (dashed) sequences to initiation of positivestrand synthesis.