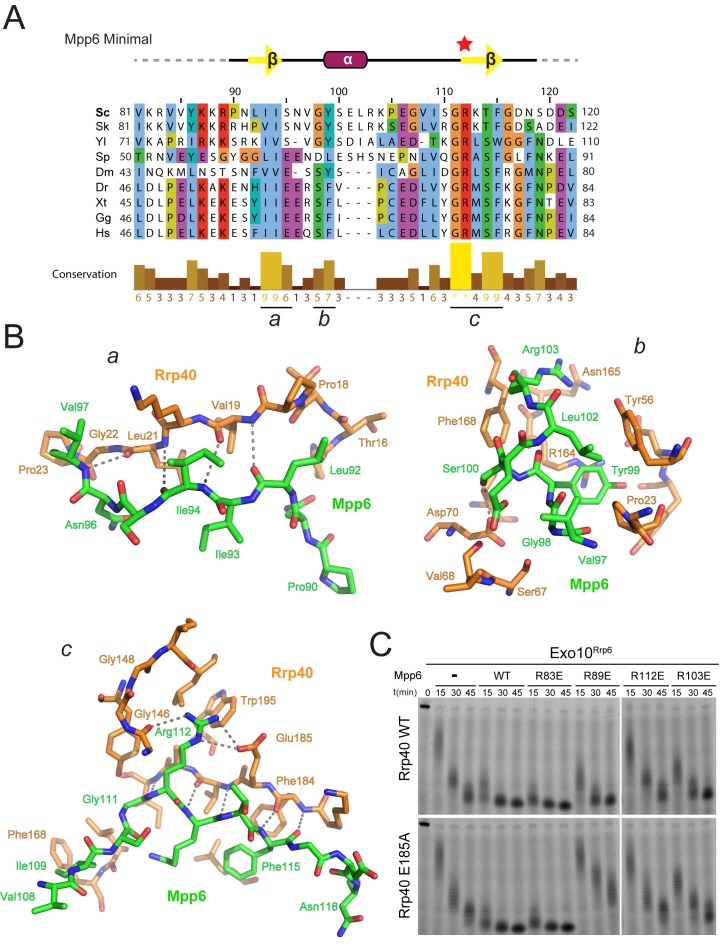
Figure 3. Conserved features of Mpp6 interaction with the S1/KH subunit, Rrp40.
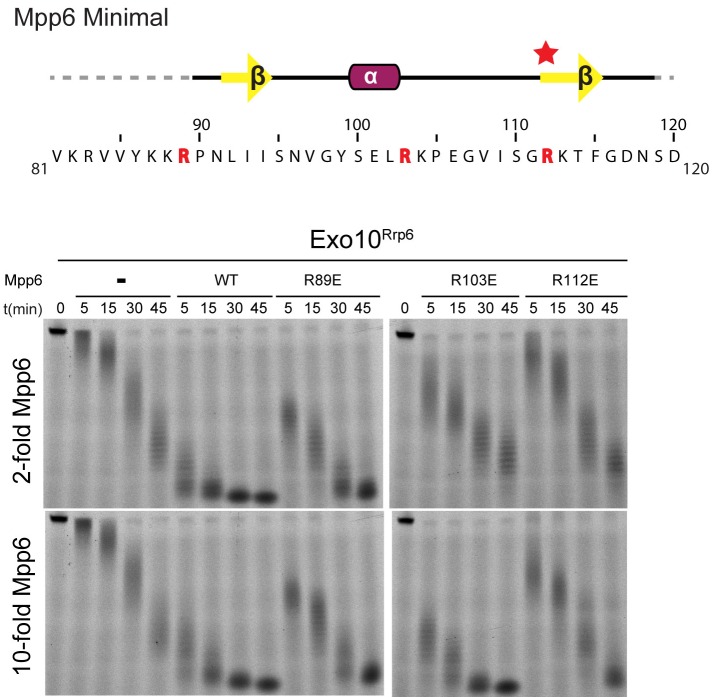
(A) Top: revised domain structure of Mpp6Minimal based on the crystal structure of Exo12Dis3exo-endo-/Rrp6exo-/Mpp6Min. Bottom: sequence alignment among S. cerevisiae Mpp6Minimal and other eukaryotes reveals conserved residues within Mpp6 Minimal. Sequences for S. kudriavzevii (Sk), Y. lipolytica (Yl), S. pombe (Sp), D. melanogaster (Dm), D. rerio (Dr), X. tropicalis (Xt), G. gallus (Gg), and H. sapiens (Hs). Sequence alignment and conservation calculated with Clustal Omega (Larkin et al., 2007) and Jalview (Waterhouse et al., 2009). Three regions (a, b, c) of high conservation are noted. (B) Stick representation of conserved contacts between Mpp6 (green) and Rrp40 (orange) are noted. Region a includes a hydrophobic patch that forms between the Rrp40 NTD and N-terminal residues of Mpp6Minimal. Region b highlights an aromatic residue in Mpp6 that nestles between the NTD and KH domains of Rrp40. Region c focuses on the network of contacts between the ‘arginine anchor’ of Mpp6 and the S1 and KH domains of Rrp40. (C) Mpp6 arginine residues were individually substituted to glutamate residues to interrogate the importance of the arginine anchor. Representative RNA degradation assays of 5’ fluorescein-labeled 49 nt polyA RNA and Exo10Rrp6 with Mpp6Minimal WT and arginine mutants. Mutations disrupting the arginine anchor, Arg112, are most detrimental to Mpp6-mediated stimulation of Rrp6 activity. An additive effect is observed when a distal Mpp6 mutation, Arg89, is combined with a mutation in Rrp40 (E185A) that disrupts coordination of the Mpp6 arginine anchor.