Fig. 1.
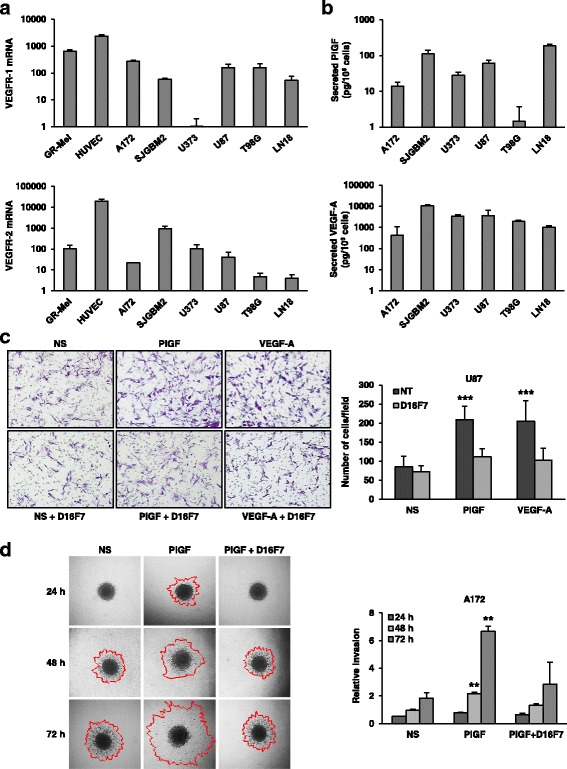
D16F7 inhibitory effects on VEGF-A or PlGF-induced migration and ECM invasion in human GBM cells expressing VEGFR-1. a Detection of VEGFR-1 and VEGFR-2 transcripts in GBM cell lines was performed by qRT-PCR. Results indicate relative mRNA expression and are the mean ± SD of three independent determinations. b PlGF and VEGF-A secretion was quantified by ELISA (mean ± SD, n = 3). c Migration of U87 cells in response to PlGF or VEGF-A was evaluated in the absence (not treated, NT) or presence of 5 μg/ml D16F7; NS, non-stimulated cells. Representative photographs of U87 cells are shown (100× magnification). Histograms represent the mean ± SD (n = 3) of migrated cells/microscopic field. Results of statistical analysis using one-way ANOVA, followed by Bonferroni’s post-test were as follows: PlGF vs NS, PlGF vs D16F7 or PlGF vs PlGF + D16F7 and VEGF-A vs NS, VEGF-A vs D16F7 or VEGF-A vs VEGF-A + D16F7, p < 0.001 (***); differences between NS, D16F7, PlGF + D16F7 or VEGF-A + D16F7 were not significant. d For spheroid invasion assay A172 cells were embedded in matrigel in the absence or presence of D16F7 (10 μg/ml) and PlGF (50 ng/ml). Representative pictures of spheroids taken at 24, 48 and 72 h after embedding cells in matrigel (40× magnification) are shown. NS, non-stimulated cells. Relative invasion was quantified as spheroid area difference (in mm2) at each of the indicated time points minus day 0. Data are expressed as mean ± SD of triplicate samples and results of statistical analysis were as follows: PlGF vs NS and PlGF vs PlGF + D16F7 at 48 and 72 h, p < 0.01 (**). Differences between NS and PlGF + D16F7 were not significant
