Figure 1. The crosstalk between metabolism and epigenetic control of gene expression in cancer.
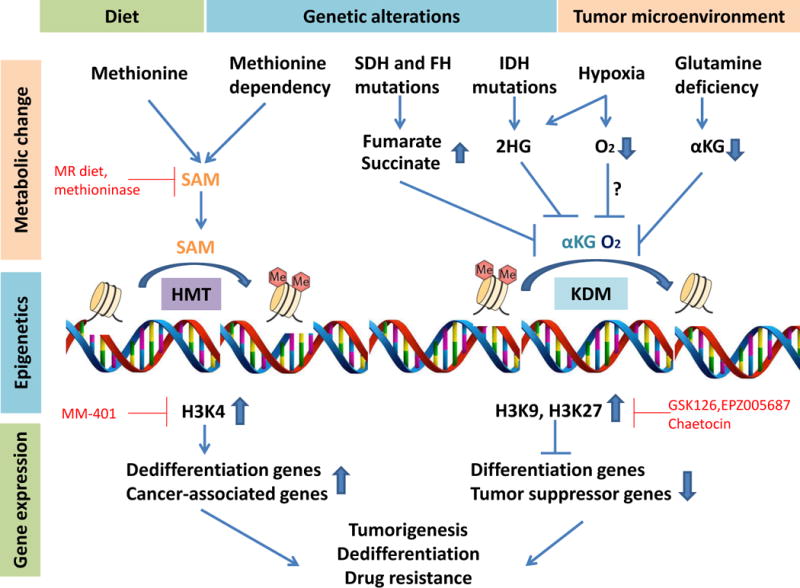
(Left) Methionine dietary intake or methionine dependency leads to increased S-adenosylmethionine (SAM) levels which promote histone methylation catalyzed by histone methyltransferase (HMT). Subsequently, hypermethylation at active marker H3K4 induces transcription of dedifferentiation genes and cancer-associated genes. (Right) Fumarate or succinate accumulation due to the loss-of-function mutant fumarate hydratase (FH) or mutant succinate dehydrogenase (SDH), 2-hydroxyglutarate (2HG) accumulation due to the mutant isocitrate dehydrogenase (IDH) or hypoxia, depletion of α-ketoglutarate (α-KG) due to glutamine deficiency, or low oxygen levels during hypoxia inhibits histone demethylation process catalyzed by histone demethylases (KDM). Subsequently, hypermethylation at suppressive histone makers H3K9 or H3K27 inhibits transcription of differentiation genes and tumor suppressor genes. Together, these metabolic regulations of epigenetics and genes expression may contribute to cell dedifferentiation, tumor progression and drug resistance in cancer. Potential therapeutic approaches to target the intersection of metabolism and gene expression in cancer are highlighted in red.
