Figure 1.
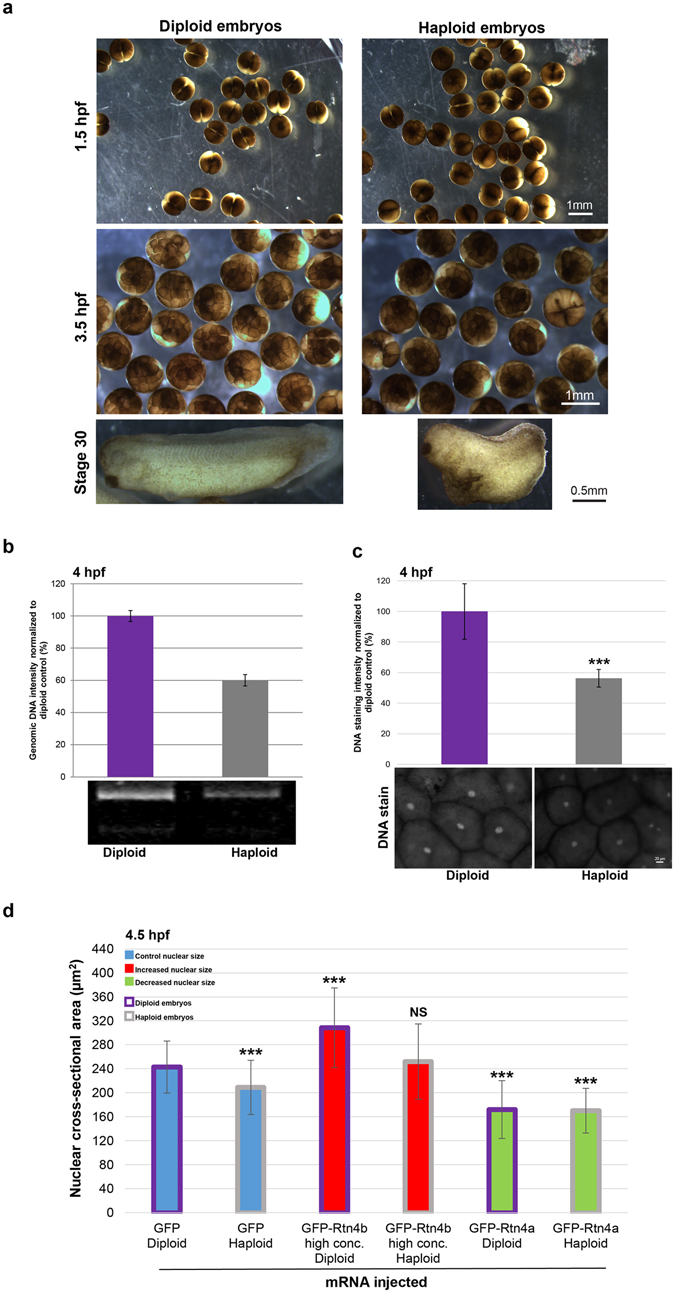
Altering ploidy in early X. laevis embryos (a) Haploid X. laevis embryos were generated by fertilizing eggs with UV-irradiated crushed X. laevis testes. Both haploid and diploid embryos underwent the first cleavage at ~1.5 hpf. Haploidy was verified by the irregular morphology of the tadpoles and their inability to develop past the swimming stage. (b) Genomic DNA was isolated from thirty 4 hpf embryos for each condition. 0.4 embryo equivalents of genomic DNA were run on 1.2% TAE agarose gels. DNA was visualized with ethidium bromide, quantified using ImageJ, and normalized to the diploid control. One representative gel and means from two independent experiments are shown. Error bars represent SD. (c) Whole un-arrested 4 hpf diploid and haploid embryos were fixed, bleached, stained for DNA with Sytox Green, and cleared. Animal pole cells were imaged for DNA intensity quantification. Several embryos from two different batches of diploid and haploid embryos were imaged. Total number of nuclei quantified: diploid, n = 38; haploid, n = 23. Error bars represent SD. ***p < 0.001. Scale bar, 20 µm. (d) Whole microinjected, un-arrested 4.5 hpf diploid and haploid embryos were fixed, bleached, stained for DNA with Sytox Green, and cleared. Animal pole cells were imaged and nuclear cross-sectional area quantified. We previously demonstrated that nuclear cross-sectional areas accurately reflect nuclear volumes6. Several embryos from two different batches of diploid and haploid embryos were imaged. Total number of nuclei quantified: GFP diploid, n = 55; GFP haploid, n = 47; GFP-Rtn4b high conc. diploid, n = 65; GFP-Rtn4b high conc. haploid, n = 49; GFP-Rtn4a diploid, n = 44; GFP-Rtn4a haploid, n = 95. The means from two independent experiments are shown. Error bars represent SD. ***p < 0.001; NS = not significant. All nuclear size differences between haploids were statistically significant by p < 0.001.
