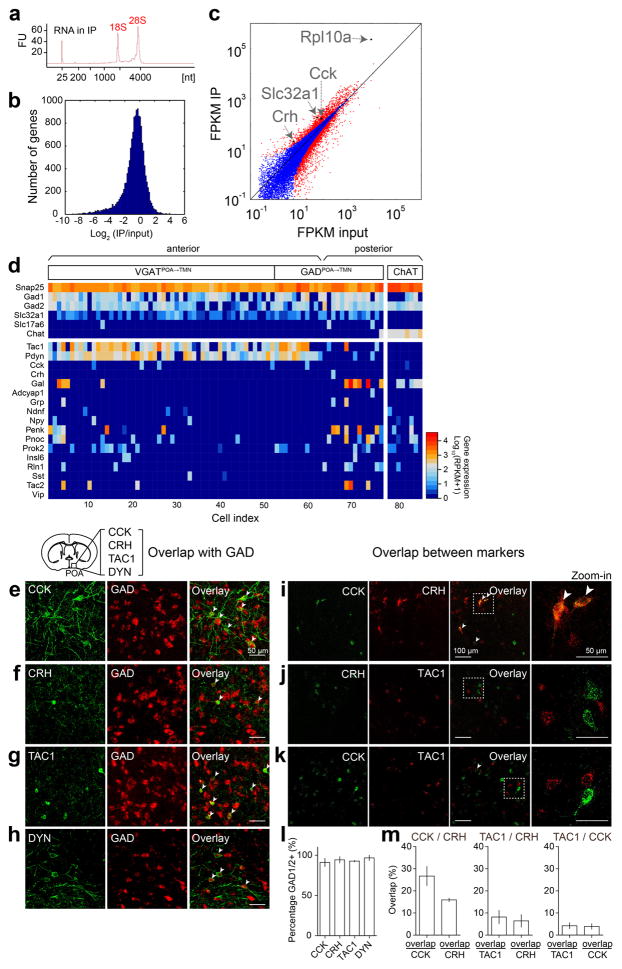
Extended Data Figure 8. Identification of genetic markers for GABAPOA→TMN neurons using TRAP and single-cell RNA-seq, and overlap between each identified marker and GAD and between the markers in the POA.
a, TRAP, shown is bioanalyzer trace of immunoprecipitated RNA. FU, fluorescence units. b, Histogram display of differentially expressed genes (IP/input). c, FPKM (fragments per kilobase of transcript per million mapped reads) IP vs. FPKM input (log scale). Several marker genes enriched in GABAPOA→TMN neurons (Cck, Crh, Slc32a1, Rpl10a) are highlighted. Red dots, genes that are significantly different in IP vs. input (P<0.05, Fisher’s exact test); blue dots, non-significant genes. d, Single-cell RNA-seq, shown is heat map of expression levels of several cell-type markers (e.g., Gad1, Gad2, Slc32a1, Slc17a6, and Chat) and all neuropeptide-encoding genes (based on the list of ref. 43 plus Gal) in cholinergic neurons in the nucleus basalis and eYFP-labeled GABAPOA→TMN neurons in the POA. Tac1 and Pdyn are highly expressed in GABAPOA→TMN neurons. RPKM, reads per kilobase of transcript per million mapped reads. e–h, Overlap between each identified marker and GAD. A representative image showing overlap between CCK-ChR2-eYFP (e), CRH-eYFP (f), TAC1-eYFP (g), DYN-ChR2-eYFP (h) and FISH for mRNA encoding GAD1/2. Arrowheads indicate cells co-labeled with GAD1/2 probe and eYFP. Mouse brain figure adapted with permission from ref. 31. i–k, Overlap between the markers. A representative image showing overlap between CCK and CRH (i), CRH and TAC1 (j) or CCK and TAC1 (k) using double FISH for both peptides. Arrowheads indicate co-labeled cells. l, Percentage of cells expressing each peptide marker that are GAD1/2 positive (n=2 or 3 mice per marker). m, Quantification of overlap between CCK and CRH, TAC1 and CRH or TAC1 and CCK (n=3 mice per pair). Error bar, ±s.e.m.