Figure 7. Model for MIR.
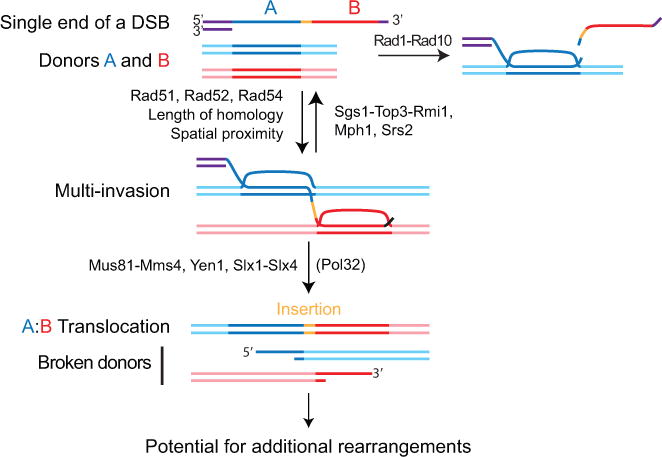
One ssDNA molecule with homology to two donors A and B, without the need for homology to one another, can form a multi-invasion intermediate. Formation of MI is stimulated by homology length and physical proximity of the donors. Cleavage of the invading strand by Rad1-Rad10 upon internal invasion irreversibly prevents MI formation and protects against MIR. The Srs2 and Mph1 (human FANCM) helicases and the Sgs1-Top3-Rmi1 (human BLM-TOPOnIα-RMI1-RMI2) helicase/topoisomerase complex disrupt MI and also inhibit MIR. Processing of MI by the overlapping activities of the Mus81-Mms4, Yen1, and Slx1-Slx4 SSE triggers MIR and transfers single-ended DSBs onto the donors. MIR occurs upon joining of the two opposite ends of the donors using the Rad51-ssDNA from the invading molecule as a synthesis template, which inserts sequence at the translocation junction. The two single-ended DSBs generated on the donors have potential to undergo secondary rearrangements. Detailed MIR mechanisms are proposed Figure S7.
