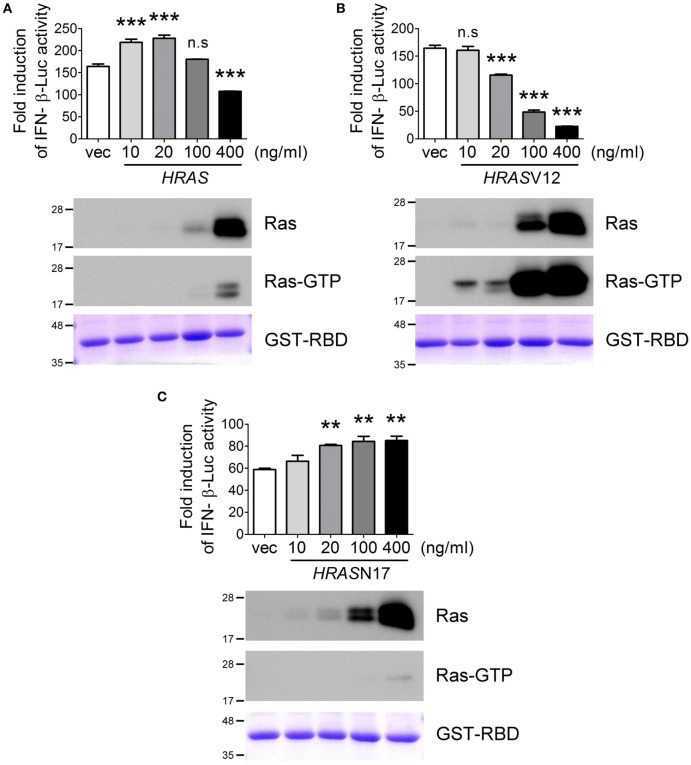
Figure 3.
The H-Ras protein enhances RLR signaling, whereas H-Ras oncogenic activity inhibits RLR signaling. HEK293T cells were transfected with the wild-type HRAS DNA (A), the HRASV12 DNA (B), or the HRASN17 DNA (C) at the indicated transfection doses together with the pIFN-β-Luc reporter plasmid and the pTK-RL reporter plasmid. In all groups, the total amounts of transfected DNA were adjusted to the same using the control vector DNA. Twenty-four hours post-transfection, cells were infected with Sendai virus (SeV) (20 HAU/ml). Luciferase activities were measured at 24 h post-infection. The results of induction fold were obtained from three independent experiments and presented as the mean ± SD. **P < 0.01 and ***P < 0.001 between the indicated groups and the control vector-transfected group (one-way ANOVA followed by Dunnett’s post hoc analysis). ns, not significant. The expression levels of transfected HRAS were detected by Western blot, shown below the bar chart. To analyze the activation status of the ectopically expressed H-Ras, we conducted another experimental set and cell lysates were harvested 24 h post SeV infection. Ras activation status was examined using GST–Ras-binding domain (RBD) Sepharose beads according to the manufacturer’s instructions. The proteins bound on the beads were separated on a 12.5% SDS-PAGE gel. The upper part of the gel (MW > 30 kDa) was stained with Coomassie blue to reveal the amount of GST–RBD precipitated (the bottommost panels), whereas the lower part of the gel (MW < 30 kDa) was processed for Western blot using a rabbit anti-H-Ras antibody to show the GTP-bound H-Ras, Ras-GTP.