Abstract
We aimed to identify whether the use of autologous hematopoietic cell transplantation (HCT) impacts outcomes for multiple myeloma patients with gains of chromosome 1q (+1q). We retrospectively identified 95 patients, 21% having +1q. For patients with +1q, the overall response rate to induction was 85%, with 40% having ≥VGPR and 20% achieving a CR, similar to non +1q patients (p =.64). The median PFS from diagnosis with +1q was 2.1 years (95% CI: 1.2–not reached (NR)) vs 4.3 years (95% CI: 3.3 yrs–NR) without +1q (p =.003). Median OS from diagnosis was 4.4 years (95% CI: 2.9–NR) vs not reached, respectively (p =.005). On molecular analysis using the Foundation One Heme assay, the most common mutations seen in +1q patients included TP53 (38%) and KRAS (25%). Overall, gain of 1q portends worse PFS and OS which was not negated by auto HCT. Such patients will likely require additional therapy to improve their survival.
Keywords: Neoplasia, myeloma, marrow and stem cell transplantation, clinical results, neoplasia–molecular genetics, neoplasia–cytogenetics
Introduction
Gains in chromosome 1 (+1q) are among the most common cytogenetic abnormalities in multiple myeloma (MM) and are identified in 20–50% of newly diagnosed patients.[1,2] The frequency increases along the disease continuum from smoldering to relapsed disease.[3] Abnormalities include balanced translocations, amplifications, and jumping translocations, leading to an increased copy number of genes on this locus.[4] Various studies have shown associations between +1q and the presence of other fluorescence-in-situ hybridization (FISH) abnormalities including del 17p and del 13q, as well as other markers of disease burden including beta-2-microglobulin, lactate dehydrogenase, anemia, bone marrow plasmacytosis, and International Staging System (ISS) stage 3 disease. Furthermore, a higher portion of patients with +1q are non-Caucasian, have IgA subtype disease, and have extramedullary or central nervous system involvement, which are associated with more aggressive disease biology.[3,5–8]
In the era of novel agent therapy for MM, cytogenetic risk stratification remains an important prognostic factor.[9,10] The International Myeloma Working Group (IMWG) defines cytogenetically high-risk disease by the presence of t(4;14) or del 17p13.[9] While gains in chromosome 1q by FISH are not currently considered high-risk by the IMWG classification, several studies have identified it as a poor prognostic marker even when patients are treated with newer active agents.[8,11,12] Other studies have failed to show a similar association, though some of these patients were treated with older regimens.[3,7,13] Studies are also conflicting on the prognostic importance of copy number variations.[8,11,14] Despite inconsistencies in available data, to be classified as low-risk by IMWG criteria, a patient cannot harbor extra copies of chromosome 1q.
While the timing of high dose melphalan and autologous hematopoietic cell transplantation (HCT) is currently being studied, upfront HCT remains standard of care for transplant-eligible patients with MM. When compared with patients without high-risk cytogenetics, MM patients with t(4;14) and del 17p have a similar depth of response, but a shorter progression free survival (PFS) duration even after HCT in the modern era.[15] In the current study, we aimed to evaluate the outcomes of patients with gains of chromosome 1 q who underwent upfront HCT.
Subjects and methods
Newly diagnosed symptomatic MM patients treated at Memorial Sloan Kettering Cancer Center (MSKCC) between January 1, 2009 and December 31, 2012 were evaluated for inclusion in our retrospective study. Patients were included if they had FISH on a pre-treatment bone marrow specimen performed as previously described,[16] received induction therapy, and subsequently underwent an upfront HCT.
Demographic and disease characteristics, as well as treatments, were abstracted from the electronic medical record with approval from the institutional review board and compared using the Fisher’s exact test. Patients were classified by the ISS stage,[17] and response was evaluated using the International Myeloma Working Group (IMWG) uniform response criteria.[18] For patients with +1q, the copy number and percentage of cells involved were collected from the cytogenetic database.
PFS and overall survival (OS) were calculated from diagnosis and HCT, estimated by Kaplan–Meier methods, and compared by the log rank test. Patients who remained alive were censored at last follow-up. Cox regression was used to create the univariate and multivariate models to examine the correlation between covariates and survival. All analyses were performed using SAS 9.4 (The SAS Institute, Cary, NC).
A subset of banked samples were analyzed using the Foundation One Heme sequencing assay, which does not require a paired normal control.[19] Using bone marrow aspirate samples, this test identifies short variants, copy number gains and losses, rearrangements, and fusions using a hybrid capture, high depth, targeted sequencing platform to analyze 405 DNA genes and 265 RNA genes.
Results
Patients
Ninety-five patients met inclusion criteria and had a median age of 58 years (range 29–73), with 63% male and 78% Caucasian. The majority of patients had IgG subtype disease (54%), and 52%, 28%, and 20% were ISS Stage I, II, and III, respectively. Fifteen percent had high-risk disease with deletion 17p or t(4;14) by FISH. Patients had a high disease burden with a median bone marrow plasmacytosis of 37% (range, 2–94%). At diagnosis, lytic lesions were identified in 79/95 (83%) with 45/95 (47%) having extramedullary disease. Creatinine above 2 mg/dL was seen in 5/95 (5%). Only one patient had central nervous system disease (Table 1).
Table 1.
Patient, disease, and treatment characteristics.
| Total sample N =95, % | Gain 1q N =20, % | No gain 1q N =75, % | p Value | |
|---|---|---|---|---|
| Age, Median (Range) | 58 (29–73) | 55 (32–71) | 58 (29–73) | .1 |
| Male | 60 (63) | 16 (80) | 44 (57) | .12 |
| Race | .89 | |||
| Caucasian | 74 (78) | 15 (75) | 59 (79) | |
| African American | 17 (18) | 4 (20) | 13 (17) | |
| Other | 4 (4) | 1 (5) | 3 (4) | |
| Isotype | .44 | |||
| IgG | 51 (54) | 14 (70) | 37 (49) | |
| IgA | 20 (21) | 3 (15) | 17 (23) | |
| Kappa | 13 (14) | 1 (5) | 12 (16) | |
| Lambda | 11 (12) | 2 (10) | 9 (12) | |
| ISS stage | >.99 | |||
| I | 49 (52) | 10 (50) | 39 (52) | |
| II | 27 (28) | 6 (30) | 21 (28) | |
| III | 19 (20) | 4 (20) | 15 (20) | |
| BM PC, % (range) | 37 (2–94) | 40 (3–82) | 36 (2–94) | .26 |
| Lytic lesions | 79 (83) | 19 (95) | 60 (80) | .18 |
| Extramedullary disease | 45 (47) | 13 (65) | 32 (43) | .085 |
| CNS disease | 1 (1) | 1 (5) | 0 | .21 |
| Creatinine >2mg/dL | 5 (5) | 1 (5) | 4 (5) | >.99 |
| Karyotype | .01 | |||
| Normal/single abnormality | 61 (64) | 8 (40) | 53 (71) | |
| Complex | 22 (23) | 10 (50) | 12 (16) | |
| Not done/not adequate | 12 (13) | 2 (10) | 10 (13) | |
| FISH | ||||
| Any abnormality | 75 (56) | 25 (100) | 50 (46) | <.001 |
| High risk | 14 (15) | 4 (20) | 10 (13) | .33 |
| 11q23 (MLL) | 29 (31) | 10 (50) | 19 (25) | .078 |
| Del13q | 25 (26) | 11 (55) | 14 (19) | .004 |
| Treatment | ||||
| Induction | .16 | |||
| Lenalidomide based | 24 (25) | 3 (15) | 21 (28) | |
| Bortezomib based | 37 (39) | 7 (35) | 30 (40) | |
| RVD | 33 (35) | 9 (45) | 24 (32) | |
| Required additional induction | 15 (16) | 3 (15) | 12 (16) | >.99 |
| Tandem HCT | 7 (7) | 1 (5) | 6 (8) | >.99 |
| Maintenance after HCT | 73 (77) | 15 (75) | 58 (77) | .78 |
| Allogeneic HCT | 14 (15) | 8 (40) | 6 (8) | .001 |
BM PC: bone marrow plasma cells; CNS: central nervous system; Del13q, deletion of chromosome 13q or monosomy 13 by FISH; FISH: fluorescence in-situ hybridization; Gain 1q: Gain of chromosome 1q21 or 1q25 by FISH; HCT: hematopoietic cell transplantation; High risk, deletion 17p, t(4;14), or t(14;16) by FISH; ISS: International Staging System; MLL: mixed lineage leukemia gene; RVD: lenalidomide, bortezomib, dexamethasone.
Patients were separated into two cohorts based on the presence or absence of a gain in chromosome 1q by FISH, with 20 patients (21%) having the abnormality. Complex karyotypes and deletion 13q by FISH were seen more frequently in patients with +1q (50 vs 16%, p =.01 and 55 vs 19%, p =.004, respectively). Lytic lesions at diagnosis and extramedullary disease were also more common in patients with excess 1q, though not statistically significant, likely due to a small sample size (95 vs 80%, p =.18 and 65 vs 43%, p =.085, respectively). The other disease characteristics were similar between the two groups (Table 1).
All 20 patients had at least 3 copies of 1q with a median of 17% (range, 1.6–98%) of cells involved. More than 3 copies were seen in 24% of patients, with a median of 48% (range, 1.6–98%) of cells involved. No patients had more than 5 copies identified by FISH.
Treatment
Induction therapy was divided into three groups for comparison. “Lenalidomide-based” included primarily lenalidomide and dexamethasone (Rd) treatment that was given to 15% of those with +1q and 28% of those without. Thirty-five percent of patients with +1q and 40% of those without +1q received “bortezomib-based” therapy, which included bortezomib and dexamethasone (Bd); cyclophosphamide, bortezomib, and dexamethasone (CyBorD); and bortezomib, liposomal doxorubicin, and dexamethasone (BDD). Finally, bortezomib, lenalidomide, and dexamethasone (RVD) was given to 45% of patients with +1q versus 32% of patients with 2 copies of 1q.
Responses to initial therapy were similar between patients with and without +1q (Table 2). For patients with +1q, the overall response rate to induction (ORR) was 85%, with 40% achieving ≥VGPR and 20% CR. Hematologic responses in patients with a normal 1q complement were not significantly different (p =.64) with an ORR of 83%, ≥VGPR 52%, and CR 20%. Patients were offered additional therapy prior to HCT if they had stable or progressive disease following initial therapy with 15% and 16% of patients with and without excess 1q requiring salvage therapy (p >.99).
Table 2.
Response to induction and auto HCT.
| Total sample N =95, % |
Gain 1q N =20, % |
No gain 1q N =75, % |
p-Value | |
|---|---|---|---|---|
| Induction | .64 | |||
| CR | 19 (20) | 4 (20) | 15 (20) | |
| VGPR | 28 (29) | 4 (20) | 24 (32) | |
| PR | 32 (34) | 9 (45) | 23 (31) | |
| SD/PD | 16 (17) | 3 (15) | 13 (17) | |
| HCT | .91 | |||
| CR | 51 (54) | 11 (55) | 40 (53) | |
| VGPR | 20 (21) | 5 (25) | 15 (20) | |
| PR | 13 (14) | 3 (15) | 10 (13) | |
| SD/PD | 3 (3) | 1 (5) | 2 (3) |
Auto HCT: autologous hematopoietic cell transplantation; CR: complete response; PD, progressive disease; PR, partial response; SD, stable disease; VGPR: very good partial response.
All patients underwent HCT with melphalan conditioning. Tandem HCT was performed in 5% of patients with +1q and in 8% without +1q (p >.99). Response rates were again similar between cohorts (p =.91) with ORR to HCT 95%, ≥VGPR 80%, and CR 55% in the +1q patients compared with 97%, 73%, and 53% in those without excess 1q, respectively. Fifteen percent of patients in each group received consolidation therapy after transplant and the vast majority received maintenance therapy (75 and 77%, p =.78), most often with lenalidomide. More patients in the +1q group went on to receive an allogeneic transplant (Allo HCT) after relapse (40% vs 8%, p =.001). Complete response rates to Allo HCT were similar in patients with +1q (63%) compared with the small number of patients without +1q (67%) (p >.99).
Survival
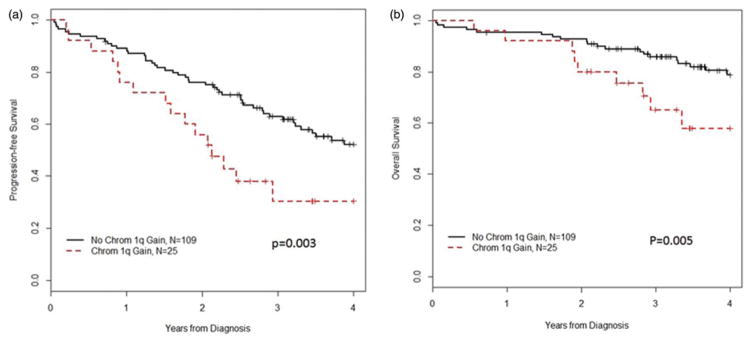
Median follow-up in surviving patients was 3.9 years (range, 0.72–6.1). The median PFS from diagnosis in patients with +1q was 2.1 years (95%CI: 1.2–not reached (NR)) compared with 4.3 years (95% CI: 3.3 yrs–NR) for patients with 2 copies of chromosome 1q (p =.003, Figure 1(a)). Similarly, two-year and three-year PFS were lower in +1q patients (56% (95% CI: 40–79%) and 31% (95% CI: 16–60%) versus 76% (95% CI: 68–84%) and 63% (95% CI: 54–73%), respectively).
Figure 1.
Survival from diagnosis, (a) progression free, (b) overall.
For +1q patients, median OS from diagnosis was 4.4 years (95% CI: 2.9–NR), with two- and three-year OS 80% (95% CI: 66–97%) and 65% (95% CI: 48–88%), respectively. Patients without +1q had a median OS that was not reached, with two- and three-year OS 93% (95% CI: 88–98%) and 86% (95% CI: 79–93%), respectively (p =.005) (Figure 1(b)).
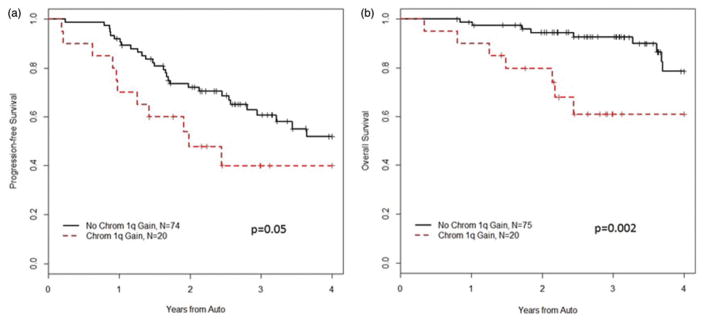
To evaluate the potential benefit of HCT, we measured PFS and OS from the date of transplantation. While the response rate of the patients with +1q remained high and similar to patients without +1q, their poor prognosis was not negated by high dose melphalan and HCT. Median PFS from HCT in patients with +1q was 1.98 years (95% CI: 1.25–NR) with two-and three-year PFS 48% (95% CI: 30–77%) and 40% (95% CI: 22–72%), respectively. On the other hand, without +1q, the median PFS was 4.4 years (95% CI: 2.96–NR) with two- and three-year PFS 72% (95% CI: 62–83%) and 61% (95% CI: 50–74%), respectively (p =.05) (Figure 2(a)). Two patients in the +1q cohort and 8 patients in the other cohort progressed prior to HCT. When these events are removed to capture only first progression following HCT, the PFS after transplantation remains shorter in patients with +1q (p =.031).
Figure 2.
Survival from auto HCT, (a) progression free, (b) overall.
Finally, the median OS from HCT was not reached in either cohort. However, two-year OS was lower in +1q patients (80% (95% CI: 64–99%) versus 94% (95% CI: 89–99%) in those without +1q. Similarly, three-year OS was 61% (95% CI: 42–89%) in patients with excess 1q compared with 93% (95% CI: 86–99%) (p =.002) (Figure 2(b)). In a comparison removing patients with del17p and t(4;14) from both cohorts, three-year OS remains shorter for the +1q patients (p =.04).
Univariate and multivariate analyses were performed to determine factors associated with PFS and OS after HCT. In the univariate model for PFS, we included the gain of chromosome 1, gender, isotype, ISS stage, and the presence of lytic lesions or extramedullary disease at diagnosis. There was a trend towards significance for the association between PFS and the presence of +1q by FISH (HR 1.99 (95% CI: 0.99–4.00), p =.05). When adjusted for ISS stage in the multivariate model, the presence of excess 1q had a significant association with PFS (HR 2.03 (1.00–4.09), p =.049) (Table 3). Including similar factors, gain of chromosome 1q was significantly associated with OS on both univariate (HR 4.6 (95% CI: 1.66–12.76), p =.003) and multivariate analysis (HR 5.11 (95% CI: 1.77–14.76), p =.004) (Table 4).
Table 3.
Univariate and multivariate analysis for PFS from auto HCT.
| Covariates | Univariate | Multivariate | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Chrom 1 gain | 1.99 (0.99–4.00) | .05 | 2.03 (1.00–4.09) | .049 |
| Male gender | 1.64 (0.88–3.05) | .12 | – | |
| Isotype – IgG | Reference | .69 | – | |
| IgA | 0.75 (0.32–1.75) | |||
| Kappa LC | 1.32 (0.59–2.98) | |||
| Lambda LC | 0.76 (0.26–2.22) | |||
| ISS stage – I | Reference | .27 | Reference | .25 |
| II | 1.5 (0.74–3.04) | 1.53 (0.76–3.12) | ||
| III | 1.85 (0.84–4.04) | 1.86 (0.85–4.08) | ||
| Lytic lesions at dx | 0.9 (0.41–1.94) | .78 | – | |
| EMD at dx | 1.54 (0.83–2.85) | .17 | – | |
Auto HCT: autologous hematopoietic cell transplantation; dx: diagnosis; EMD: extramedullary disease; ISS: International Staging System; LC: light chain; PFS: progression-free survival. Bold value indicates significant p values.
Table 4.
Univariate and multivariate analysis for OS from auto HCT.
| Covariates | Univariate | Multivariate | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Chrom 1 gain | 4.6 (1.66–12.76) | .003 | 5.11 (1.77–14.76) | .004 |
| Male gender | 1.4 (0.53–3.68) | .5 | – | |
| Isotype – IgG | Reference | .75 | – | |
| IgA | 0.66 (0.18–2.38) | |||
| Kappa LC | 0.61 (0.13–2.75) | |||
| Lambda LC | 0.44 (0.06–3.45) | |||
| ISS stage – I | Reference | .16 | Reference | .14 |
| II | 2.74 (0.9–8.4) | 2.98 (0.96–9.19) | ||
| III | 2.45 (0.66–9.16) | 2.73 (0.72–10.26) | ||
| Lytic lesions at dx | 0.72 (0.23–2.2) | .56 | – | |
| EMD at dx | 2.46 (0.91–6.67) | .08 | – | |
Auto HCT: autologous hematopoietic cell transplantation; dx: diagnosis; EMD: extramedullary disease; ISS: International Staging System; LC: light chain; PFS: progression-free survival. Bold values indicate significant p values.
Molecular
The Foundation One Heme assay was used to identify genomic abnormalities in a subset of patients with available banked samples. Twenty-nine samples were examined, with 8 samples from +1q patients. These included 3 diagnostic samples and 5 relapse samples, all of which had complex karyotypes. The other 21 samples were divided as follows: diagnostic samples with normal cytogenetics and karyotype (n =8); diagnostic samples with normal cytogenetics and karyotype, but primary refractory disease (n =2); diagnostic samples with other cytogenetic abnormalities, but primary refractory disease (n =4); relapse samples of patients with primary refractory disease (n =5); samples with complex karyotypes, but no +1q (n =2).
The median exon coverage was 494.5 × (range 381–603× per sample), and the median percentage of reads supporting the variant was 48 (range 1–100). Of note, 13 patients’ results (45%) were qualified due to alteration detection sensitivity reduced from low coverage, elevated GC bias, low tumor content, or potential contamination.
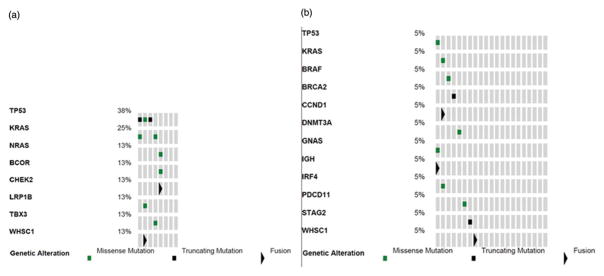
A total of 267 genomic alterations were identified, with 239 (90%) classified as variants of unknown significance. For descriptive purposes, we will discuss the known and likely somatic alterations (Figure 3). In the +1q patients, 3/8 patients had more than one alteration with the most common mutations being TP53 (38%) and KRAS (25%). In the patients without +1q, a broader range of mutations was seen with all mutations found once in the cohort, with 1 mutated TP53 and 1 KRAS. Interestingly, 3/8 patients with +1q and 13/21 patients without +1q had no known or likely variants.
Figure 3.
Genomic abnormalities called by Foundation One as known or likely, (a) Patients with +1q by FISH, (b) Patients without +1q by FISH.
Finally, CKS1B is one of the genes proposed to be amplified with a gain in 1q and has been associated with poor prognosis. None of the eight +1q samples had amplifications of CKS1B. However, 7 copies are required to be considered amplified by the Foundation One Heme assay. On subsequent analysis of the data, one patient had 7 copies, but the sample was qualified for low purity. Four of the patients with-+1q by FISH had 3–4 copies identified. Intriguingly, two patients without +1q by FISH were also identified as having 3 copies of CKS1B.
Discussion
We present a large retrospective study evaluating the disease characteristics and prognosis of patients with gains of chromosome 1q by FISH who were treated with novel agent induction regimens and high-dose melphalan with HCT. While response rates to induction were high and similar to patients without these cytogenetic abnormalities, patients with +1q had shorter PFS and OS from diagnosis. Furthermore, the poor prognosis conferred by +1q was not abrogated by HCT.
Recent studies have evaluated the outcomes of patients with +1q based on induction regimen. An et al. [11] showed that patients with +1q had inferior outcomes when treated with bortezomib, but not thalidomide. They also showed that three copies of 1q21 in 20% of plasma cells conferred bortezomib resistance. Similarly, Biran et al. [8] showed that these patients had a higher frequency of markers of aggressive disease and a median OS from diagnosis of only 37 months when treated with RVD induction. Our results are similar and confirm that patients with +1q have a poorer OS from diagnosis regardless of initial therapy.
More recently, Kazmi et al. [15] demonstrated that patients with high-risk cytogenetic abnormalities have a worse prognosis after HCT compared with standard-risk patients. They used a broader definition of high-risk and did not separate the results based on individual abnormalities. However, they showed that these patients have prolonged survival if transplanted in first remission rather than at relapse. On the other hand, in the first interim analysis of a phase III trial evaluating HCT vs bortezomib-based consolidation after a bortezomib based induction, Cavo et al. [20] found a longer PFS with HCT and that this benefit was retained across pre-defined subgroups including high-risk disease defined as t(4;14) ± del(17p) ± del(1p) ± 1q gain. Further information on the number of patients in these subgroups and their outcomes compared with those transplanted without the abnormalities will be important. In a retrospective analysis of the +1q subset presented by Bock et al.,[21] median PFS from transplant was only 10.7 months. With longer follow-up, we are able to show that beyond just a shorter median PFS, patients with +1q also have a shorter OS from HCT. However, as we only included patients with upfront HCT, we are unable to compare the outcomes to those transplanted at relapse.
Several genes have been suggested as the target on chromosome 1q. By the 70-gene molecular signature developed at Arkansas (GEP70), 30% of high-risk genes are on chromosome 1.[22] Two particular proposed genes are CKS1B, which regulates ubiquitination and proteolysis of cyclin-dependent kinases to mediate cell cycle progression and confers drug resistance, and PMSD4, a polyubiquitin receptor that mediates bortezomib resistance.[23,24] In our subset of banked samples, we found an increase in copy number of CKS1B, though not to the level required by clinical sequencing assays. However, as copy number variation is not uniformly associated with prognosis, additional data on CD138 selected samples will likely allow for a more robust analysis. In addition, studies are underway to identify agents that can overcome CKS1B-induced drug resistance.[25]
In our limited sample, we also found that additional molecular abnormalities may be more common in the +1q patients, potentially allowing for the segregation of these patients into subgroups for targeted therapies. TP53 and KRAS mutations were seen in +1q patients, but additional analysis in a larger set of patients would be needed to confirm this finding. Walker et al. [26] identified mutations in the RAS/MAPK, nuclear factor-κB, and DNA repair pathways as the most common in newly diagnosed MM, though the prognostic significance varies. In their samples of all newly diagnosed patients, RAS mutations occurred in 43% of samples, while TP53 pathway alterations occurred in 11%. Similarly, using deep sequencing, Lionetti et al. [27] found that more than half the patients had alterations in the MAPK pathway, but no data with respect to therapy was given. Therefore, the implications of such mutations in the context of HCT remains unclear.[28] These studies suggest that studying agents that target the MAPK pathway may be effective for treating MM including patients with excess 1q.
Additional options for therapy intensification for patients with +1q may include the use of tandem HCT frontline or modifications to the high dose melphalan conditioning regimen. Recent studies of modern induction including proteasome inhibition followed by tandem HCT in patients with high-risk disease have shown three-year PFS between 35 and 69%.[14,29] In addition, studies adding bendamustine or thiotepa to melphalan as conditioning for the second HCT suggest that these options are feasible,[30,31] though their impact is unknown as only 10% of patients in each trial had high-risk disease by current cytogenetic risk classification. Pharmacokinetic targeted dosing of melphalan may also improve outcomes while limiting toxicity.[32]
There are several limitations of this study, including its retrospective nature. In addition, during the included time period, standard FISH was performed on unselected bone marrow aspirates. FISH performed on CD138 selected cells, as is our practice now, is significantly more sensitive and allows for better cytogenetic characterization. Our pathology department also used probes for 1q25 and 1q21 depending on the lab standard at the time. This is unlikely to affect the results as the entire arm is often duplicated by karyotype. Furthermore, we had planned to compare copy number alterations with outcome, but most patients had 3–4 copies by FISH, and therefore, this analysis was not possible. Similarly, as the Foundation One Heme requires 7 copies for a gene to considered amplified, we were unable to molecularly define our population based on these limited samples. However, we intend to expand our studies to include more recently evaluated patients with CD138 selected samples to confirm our findings.
In conclusion, patients with gains in chromosome 1 have a worse PFS and OS even after undergoing HCT. Additional studies are needed to identify treatments that can overcome these resistant MM cell clones. Molecular sequencing may identify potential targets but will require study in a large population of +1q patients. Finally, additional interventions such as newer conditioning regimens, tandem transplants, or molecularly targeted therapies from induction and continued throughout therapy may be needed to improve outcomes.
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article at http://dx.doi.org/10.1080/10428194.2016.1260126.
References
- 1.Liebisch P, Wendl C, Wellmann A, et al. High incidence of trisomies 1q, 9q, and 11q in multiple myeloma: results from a comprehensive molecular cytogenetic analysis. Leukemia. 2003;17:2535–2537. doi: 10.1038/sj.leu.2403153. [DOI] [PubMed] [Google Scholar]
- 2.Sawyer JR, Waldron Ja, Jagannath S, et al. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995;82:41–49. doi: 10.1016/0165-4608(94)00284-i. [DOI] [PubMed] [Google Scholar]
- 3.Hanamura I, Stewart JP, Huang Y, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marzin Y, Jamet D, Douet-Guilbert N, et al. Chromosome 1 abnormalities in multiple myeloma. Anticancer Res. 2006;26:953–959. [Google Scholar]
- 5.Wu KL, Beverloo B, Lokhorst HM, et al. Abnormalities of chromosome 1p/q are highly associated with chromosome 13/13q deletions and are an adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Br J Haematol. 2007;136:615–623. doi: 10.1111/j.1365-2141.2006.06481.x. [DOI] [PubMed] [Google Scholar]
- 6.Bang S-M, Kim YR, Cho HI, et al. Identification of 13q deletion, trisomy 1q, and IgH rearrangement as the most frequent chromosomal changes found in Korean patients with multiple myeloma. Cancer Genet Cytogenet. 2006;168:124–132. doi: 10.1016/j.cancergencyto.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Fonseca R, Van Wier Sa, Chng WJ, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–2040. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 8.Biran N, Malhotra J, Bagiella E, et al. Patients with newly diagnosed multiple myeloma and chromosome 1 amplification have poor outcomes despite the use of novel triplet regimens. Am J Hematol. 2014;89:616–620. doi: 10.1002/ajh.23705. [DOI] [PubMed] [Google Scholar]
- 9.Chng WJ, Dispenzieri A, Chim C-S, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28:269–277. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 10.Palumbo A, Avet-Loiseau H, Oliva S, et al. Revised international staging system for multiple myeloma: a report From International Myeloma Working Group. J Clin Oncol. 2015;33:2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An G, Xu Y, Shi L, et al. Chromosome 1q21 gains confer inferior outcomes in multiple myeloma treated with bortezomib but copy number variation and percentage of plasma cells involved have no additional prognostic value. Haematologica. 2014;99:353–359. doi: 10.3324/haematol.2013.088211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waheed S, Shaughnessy JD, van Rhee F, et al. International staging system and metaphase cytogenetic abnormalities in the era of gene expression profiling data in multiple myeloma treated with total therapy 2 and 3 protocols. Cancer. 2011;117:1001–1009. doi: 10.1002/cncr.25535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaughnessy JD, Haessler J, van Rhee F, et al. Testing standard and genetic parameters in 220 patients with multiple myeloma with complete data sets: superiority of molecular genetics. Br J Haematol. 2007;137:530–536. doi: 10.1111/j.1365-2141.2007.06586.x. [DOI] [PubMed] [Google Scholar]
- 14.Neben K, Lokhorst HM, Jauch A, et al. Administration of bortezomib before and after autologous stem cell transplantation improves outcome in multiple myeloma patients with deletion 17p. Blood. 2012;119:940–948. doi: 10.1182/blood-2011-09-379164. [DOI] [PubMed] [Google Scholar]
- 15.Kazmi SM, Nusrat M, Gunaydin H, et al. Outcomes among high-risk and standard-risk multiple myeloma patients treated with high-dose chemotherapy and autologous hematopoietic stem-cell transplantation. Clin Lymphoma Myeloma Leuk. 2015;15:687–693. doi: 10.1016/j.clml.2015.07.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerritsen WR, Donohue J, Bauman J, et al. Clonal analysis of myelodysplastic syndrome: monosomy 7 is expressed in the myeloid lineage, but not in the lymphoid lineage as detected by fluorescent in situ hybridization. Blood. 1992;80:217–224. [PubMed] [Google Scholar]
- 17.Greipp PR, San Miguel J, Durie BGM, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23:3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 18.Durie BGM, Harousseau J-L, Miguel JS, et al. International uniform response criteria for multiple myeloma. Leukemia. 2006;20:1467–1473. doi: 10.1038/sj.leu.2404284. [DOI] [PubMed] [Google Scholar]
- 19.Foundation Medicine. Foundation One Heme Technical Information and Test Overview [Internet] 2015 [cited 2015 Dec 23]. Available from: http://foun-dationone.com/docs/FM_TechnicalOverview_HEM-I-001-20150105.pdf.
- 20.Cavo M, Palumbo A, Zweegman S, et al. Upfront autologous stem cell transplantation (ASCT) versus novel agent-based therapy for multiple myeloma (MM): a randomized phase 3 study of the European Myeloma Network (EMN02/HO95 MM trial) J Clin Oncol. 2016;34:8000. [Google Scholar]
- 21.Bock FM, Lu G, Shah N, et al. Clinical characteristics and outcomes in patients with multiple myeloma and CKS1B gene gain/amplification after autologous stem cell transplantation. Blood. 2014;124:2521. [Google Scholar]
- 22.Shaughnessy JD, Jr, Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 23.Shaughnessy JD, Qu P, Usmani S, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118:3512–3524. doi: 10.1182/blood-2010-12-328252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhan F, Colla S, Wu X, et al. CKS1B, overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms. Blood. 2007;109:4995–5001. doi: 10.1182/blood-2006-07-038703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Zhou Y, Thomas GS, et al. NEDD8 inhibition overcomes CKS1B-induced drug resistance by upregulation of p21 in multiple myeloma. Clin Cancer Res. 2015;21:5532–5542. doi: 10.1158/1078-0432.CCR-15-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker BA, Boyle EM, Wardell CP, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lionetti M, Barbieri M, Todoerti K, et al. Molecular spectrum of BRAF, NRAS and KRAS gene mutations in plasma cell dyscrasias: implication for MEK-ERK pathway activation. Oncotarget. 2015;6:24205–24217. doi: 10.18632/oncotarget.4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gebauer N, Biersack H, Czerwinska A-C, et al. Single nucleotide polymorphisms in TP53 but not KRAS or MDM2 are predictive of clinical outcome in multiple myeloma treated with high-dose melphalan and autologous stem cell support. Leuk Lymphoma. 2016;57:1482–1486. doi: 10.3109/10428194.2015.1099648. [DOI] [PubMed] [Google Scholar]
- 29.Cavo M, Tacchetti P, Patriarca F, et al. Bortezomib with thalidomide plus dexamethasone compared with thalidomide plus dexamethasone as induction therapy before, and consolidation therapy after, double autologous stem-cell transplantation in newly diagnosed multiple myeloma: a randomised phase 3 study. Lancet. 2010;376:2075–2085. doi: 10.1016/S0140-6736(10)61424-9. [DOI] [PubMed] [Google Scholar]
- 30.Musso M, Messina G, Marcacci G, et al. High-dose melphalan plus thiotepa as conditioning regimen before second autologous stem cell transplantation for “De Novo” multiple myeloma patients: a phase II study. Biol Blood Marrow Transplant. 2015;21:1932–1938. doi: 10.1016/j.bbmt.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Martino M, Tripepi G, Messina G, et al. A phase II, single-arm, prospective study of bendamustine plus melphalan conditioning for second autologous stem cell transplantation in de novo multiple myeloma patients through a tandem transplant strategy. Bone Marrow Transplant. 2016;51:1197–1203. doi: 10.1038/bmt.2016.94. [DOI] [PubMed] [Google Scholar]
- 32.Nath CE, Trotman J, Tiley C, et al. High melphalan exposure is associated with improved overall survival in patients receiving high dose melphalan and autologous transplantation for myeloma. Leuk Lymphoma. 2016;82:149–159. doi: 10.1111/bcp.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]