Figure 1.
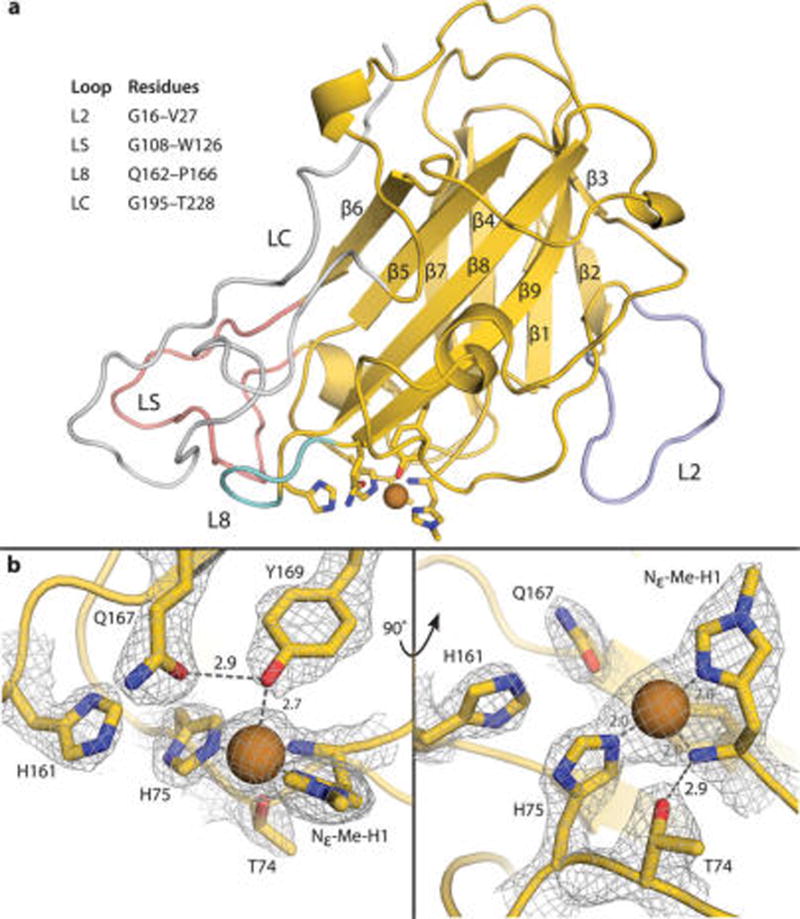
(a) Crystal structure of MtPMO3* in cartoon representation with primary and secondary coordination sphere residues as sticks. All β-strands are numbered in order of primary sequence. Relevant loop features are highlighted in lavender (L2), pink (LS), teal (L8), and silver (LC). All other carbon atoms are shown in yellow, nitrogen atoms in blue, and oxygen atoms in red. Copper ions are depicted as a brown sphere. (b) MtPMO3* active site showing primary (Nε-Me-H1, H75, Y169) and secondary (T74, H161, Q167) sphere residues with mesh electron density map contoured to 1.5 σ. H161 and Q167 are positioned to H-bond with ligands in the solvent-facing equatorial position. Q167 H-bonds with the axial tyrosine ligand. T74 H-bonds with the N-terminal amino group of H1 and forms a bond with an axial solvent ligand when present. Hydrogen-bonding distances are shown in Ångstroms (Å).
