Figure 2.
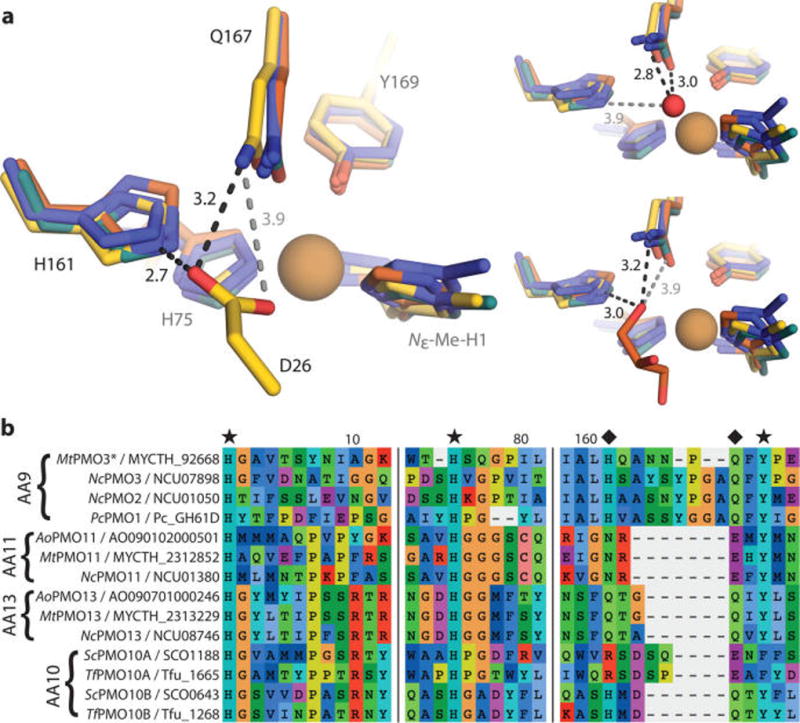
(a) Structural superposition of MtPMO3* (yellow) with three other AA9 structures, PcPMO1 (orange, PDB ID 4B5Q), NcPMO2 (indigo, 4EIR), and NcPMO3 (teal, 4EIS). Conserved residues H161 and Q167 (MtPMO3* numbering) form H-bonds with D26 from a symmetry-related molecule in MtPMO3* (left), a water molecule in NcPMO3 (top right), and a buffer-derived glycerol molecule in PcPMO1 (bottom right). Distances are in Ångstroms (Å). The active-site Cu appears reduced by the X-ray beam in all structures; exogenous molecules depicted in the solvent-facing equatorial position are at lengths (3.6–4.6 Å) from the Cu center that exclude formal coordination. (b) Multiple sequence alignment of representative AA9, AA10, AA11, and AA13 PMOs showing conservation of H-bonding motifs (marked with diamonds) within the PMO superfamily. Fungal PMOs possess a H-X5–8-Q-X-Y (AA9), N-X-E-X-Y (AA11), or Q-X2-Q-X-Y (AA13) motif depending on substrate specificity (cellulose, chitin, and starch, respectively). Bacterial AA10 PMOs active on cellulose contain either a R-X4-E-X-F or H-X2-Q-X-Y motif, correlating with observed regioselectivity (C1 or C1/C4, respectively). Primary coordinating residues are marked with a star. Numbers correspond to MtPMO3* amino acid numbering.
