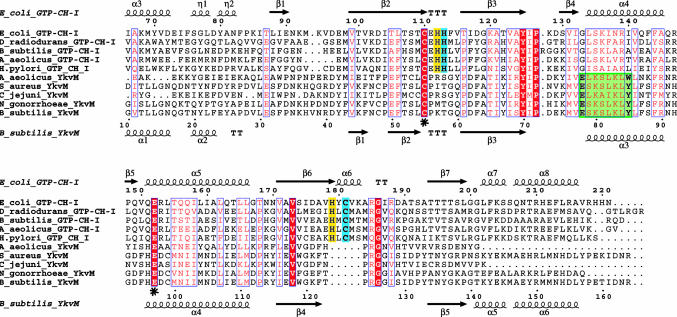
Fig. 5.
Alignment of unimodular FolE (GTP cyclohydrolase I) and YkvM sequences. For clarity and space, only sequences from select organisms are shown from among 60 sequences in the original alignment, and the N-termini have been truncated. Sequence numbers of every 10th residue are shown for E. coli FolE and B. subtilis YkvM. Secondary structure elements and nomenclature as defined by the crystal structure of E. coli FolE and by the 3D homology model of B. subtilis YkvM are shown at Upper and Lower, respectively. The conserved Cys and Glu found in the substrate binding pocket of both protein families are indicated by asterisks. The QueF motif, specific for the QueF family, is highlighted in green. The zinc binding His and Cys residues found in FolE and not in QueF are highlighted in blue. Other catalytic residues in FolE not found in QueF are highlighted in yellow. The absence of the zinc-binding and catalytic residues of FolE is the best identifier of QueF sequences in genome databases.