Summary
Fluorescent reporter and epitope tagged human pluripotent stem cells (hPSCs) greatly facilitate studies on the pluripotency and differentiation characteristics of these cells. Unfortunately traditional procedures to generate such lines are hampered by a low targeting efficiency that necessitates a lengthy process of selection followed by the removal of the selection cassette. Here we describe a procedure to generate fluorescent reporter and epitope tagged hPSCs in an efficient one-step process using the CRISPR/Cas technology. Although the method described uses our recently developed iCRISPR platform, the protocols can be adapted for general use with CRISPR/Cas or other engineered nucleases. The transfection procedures described could also be used for additional applications, such as over-expression or lineage tracing studies.
Keywords: human pluripotent stem cells (hPSCs), gene targeting, CRISPR/Cas, homologous recombination, knockin, gene targeting, fluorescent reporter, epitope tag
1. Introduction
Human pluripotent stem cells (hPSCs), which include embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs), have two unique characteristics: the capacity for unlimited self-renewal in culture and the ability, called pluripotency, to form any cell type that is present in an adult human. Since the isolation of hESCs in 1998, there has been much progress in the development of protocols to differentiate hPSCs into specific cell types. As a result hPSCs have emerged as a valuable tool for studying human development and for modeling human disease. Furthermore there is great potential to use hPSCs to generate clinically important cell types, which can be transplanted into patients to replace those cells lost or damaged due to disease (Tabar and Studer, 2014; Zhu and Huangfu, 2013).
The utility of hPSCs can be further enhanced through genetic modification of these lines. Through homologous recombination exogenous sequences can be inserted into the genome to generate a variety of “knockin” alleles, from a single nucleotide change to introduce or correct a disease associated mutation, to the insertion of small (less than 100 base pairs, bp) epitope tags (e.g. hemagglutinin, 3xFLAG, V5 tags) or even the insertion of large (hundreds of bp) transgene constructs such as fluorescent reporters (e.g. GFP, mOrange) and antibiotic selection cassettes. The ability to insert epitope or fluorescent tags into the genome would greatly facilitate hPSC-based in vitro and in vivo (after transplantation in animal models) studies. The insertion of epitope tags into specific loci creates recombinant hybrids that contain a polypeptide affinity tag and enable efficient purification of target proteins. The development of these fusion proteins facilitates a variety of downstream experiments including: western blot, immunoprecipitation (IP), chromatin-immunoprecipitation (ChIP) and immunofluorescence, and is particularly useful when antibodies for the protein of interest are substandard or not available. Lineage-specific knockin fluorescent reporters are valuable tools because they allow real-time observation of gene expression dynamics, cell lineage tracing and the isolation of a specific cell population for further analysis. hPSCs knockin reporter lines have already been used to identify mediators of pluripotency (Chia et al., 2010) as well as characterize the culture conditions that stabilize different pluripotent states (Theunissen et al., 2014).
Creating knockin alleles in hPSCs used to be extremely challenging because of the low transfection efficiency (Eiges et al., 2001) and low rate of spontaneous homologous recombination (Zwaka and Thomson, 2003). Thus traditional knockin strategies rely on the use of a drug-resistance cassette that allows for the enrichment of cells with the correct integration. However, due to potential interference between the drug-resistance cassette and nearby genes, the selection cassette must be removed prior to using these cells for experimental studies (Davis et al., 2008). We have ourselves generated a knockin reporter allele at the OCT4 locus using antibiotic selection (Zhu et al., 2015). Notably correctly targeted clones with the drug selection cassette did not express GFP. Only upon Cre-mediated deletion of the drug selection cassette, did all clonal lines show proper co-expression of pan-cellular eGFP with OCT4. Thus the low targeting efficiency of hPSCs necessitates an antibiotic selection cassette, but this in turn requires additional targeting, isolation and characterization of clonal lines, effectively doubling the time and effort required to make the knockin line.
The recent emergence of programmable site-specific nucleases has greatly improved the efficiency of genome engineering in hPSCs. Zinc finger nucleases (ZFNs), transcription-activator like endonucleases (TALENs) and the CRISPR/Cas9 nuclease can all act as genomic scissors to produce a double strand break (DSB) at the desired genome loci (Joung and Sander, 2013; Ran et al., 2013; Urnov et al., 2010). Without a repair template, the DSB is repaired by the non-homologous end joining (NHEJ) pathway that often generate small insertions or deletions (indels). These hPSC lines with site-specific knockout mutations can be used to determine the contribution of particular genes for development and disease. In the presence of a repair template, DSB repair with error-free homology directed repair (HDR) can be used to generate knockin alleles through targeted integration of exogenous DNA sequences into the desired loci.
The CRISPR/Cas9 system was discovered as a form of adaptive immunity in bacteria, where it is used to destroy exogenous nucleic acids of invading viruses (Jinek et al., 2012), and it has recently been adapted for genome engineering in mammalian cells. In this system the CRISPR chimeric guide RNA (gRNA) recognizes a 20-nucleotide (nt) DNA sequence upstream of the 5’-NGG-3’ protospacer adjacent motif (PAM) and directs the DNA endonuclease Cas9 for site-specific cleavage (Cho et al., 2013; Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013). Cas9 produces a DNA DSB, which in the presence of a double stranded DNAs (dsDNA) plasmid or single stranded DNAs (ssDNA) template can promote HDR to efficiently incorporate exogenous sequences, such as a fluorescent reporter tag, into the specific genomic locus in hPSCs (Byrne et al., 2015; Hou et al., 2013; Merkert et al., 2014; Merkle et al., 2015)(Zhu et al., 2015). We have developed an efficient genome-editing platform in hPSCs, called iCRISPR (Gonzalez et al., 2014; Zhu et al., 2014). Through TALEN-mediated gene targeting, a doxycycline inducible Cas9 cassette and the reverse tetracycline transactivator (M2rtTA) are integrated into both alleles of the endogenous AAVS1 locus, and thus allow robust Cas9 expression in established clonal lines (referred to as iCas9 hPSCs) upon doxycycline treatment. With transfection of appropriate gRNAs we are able to generate site-specific knockout mutations. In addition, with co-transfection of repair templates, iCRISPR has also allowed us to specifically alter the sequence of a small number of nucleotides, and to insert a knockin epitope tag or a large transgenes such as a fluorescent tag. Most importantly due to the increased efficiency of the iCRISPR system it is not necessary to use selection to enrich for correctly integrated alleles. Thus it is feasible to generate knockin alleles in an efficient one-step procedure (Zhu et al., 2015).
In this methods paper we describe the procedure for using our iCRISPR system to generate knockin alleles in hPSCs. We specifically describe the procedure for generating epitope tagged and fluorescent tagged hPSC lines; however, the same knockin strategy could be used for a variety of other inserts. Furthermore we believe the transfection approach that we use to generate the lines could also be used for other purposes, for example to deliver mRNA or plasmids for overexpression or lineage tracing studies.
2. Materials
hPSC medium: DMEM/F12 medium (Life Technologies) , 20% KnockOut Serum Replacement (Life Technologies), 1X Non-Essential Amino Acids (Life Technologies), 1X GlutaMAX (Life Technologies), 100 U/mL Penicillin/100 µg/mL Streptomycin (Life Technologies), 0.055 mM 2-mercaptoethanol (Life Technologies, 10 ng/mL recombinant human basic FGF (Life Technologies).
TrypLE Select enzyme (Life Technologies)
Fetal Bovine Serum (FBS, Sigma Aldrich)
Dimethyl Sulfoxide (DMSO, Santa Cruz Biotechnology)
Rho-associated protein kinase (ROCK) inhibitor Y-27632 dihydrochloride (Selleck Chemicals)
Genomic DNA lysis buffer: 10 mM Tris pH8, 0.45% NP40, 0.45% Tween 20 and 100 µg/ml Proteinase K.
Lipofectamine RNAiMAX (Life Technologies)
Lipofectamine 3000 (Life Technologies)
Herculase II Fusion DNA Polymerase (Agilent Technologies)
MEGAshortscript T7 Transcription kit (Life Technologies)
MEGAclear Transcription Clean-Up Kit (Life Technologies)
DNeasy Blood & Tissue Kit (Qiagen)
Ethidium Bromide (EtBr)
Agarose (Invitrogen)
Purelink Gel Purification Kit (Life Technologies)
Pierce Crosslink Magnetic IP/Co-IP kit (Pierce Antibodies)
FLAG antibody, clone M2 (Sigma Aldrich)
TBST: 20 mM Tris-HCl (pH 7.6), 137 mM NaCl, 0.1 % Tween 20
Blocking Buffer: 5% non-fat milk in TBST
NuPAGE LDS Sample Buffer 4x (Life Technologies)
NuPAGE® Sample Reducing Agent 10x (Life Technologies)
NuPAGE Novex 3–8% Tris-Acetate Protein Gels (Life Technologies)
NuPAGE Tris-Acetate SDS Running Buffer (20X) (Life Technologies)
Nitrocellulose Pre-Cut Blotting Membranes (Life Technologies)
Amersham ECL Prime Western Blotting Detection Reagent (Amersham)
Amersham Hyperfilm (Amersham)
Amersham Hypercassette (Fisher)
3. Methods
Here we describe the procedures to generate epitope and reporter tagged hPSC lines using the iCRSIPR platform. In the iCRISPR system a doxycycline inducible Cas9 cassette has been targeted into the endogenous AAVS1 locus. Upon doxycycline treatment, Cas9 is expressed in all cells. After transfection of the site-specific gRNAs and the donor HDR template with the exogenous DNA sequence flanked by the homology arms, a DSB is created and HDR leads to incorporation of the insert into the genome. For single nucleotide alternations, a ssDNA template with homology arms of ~ 40–80 nt are generally used. Due to the small size of the epitope tags (less than 100 nt), the exogenous sequence can also be provided as a ssDNA, whereas the larger reporter tag (hundreds of bp) can be cloned easily into a donor plasmid for delivery into the cells. In both cases selection is not required and knockin alleles can be efficiently generated in a single step (Fig. 1). We have used the following protocols for HUES8 (NIHhESC-09–0021) and MEL-1 (NIHhESC-11–0139) hESC lines. It may be necessary to adjust the protocols for different hPSC lines or for hPSCs cultured under different conditions such as feeder-free TeSR and Essential 8 culture conditions (Chen et al., 2011; Ludwig et al., 2006).
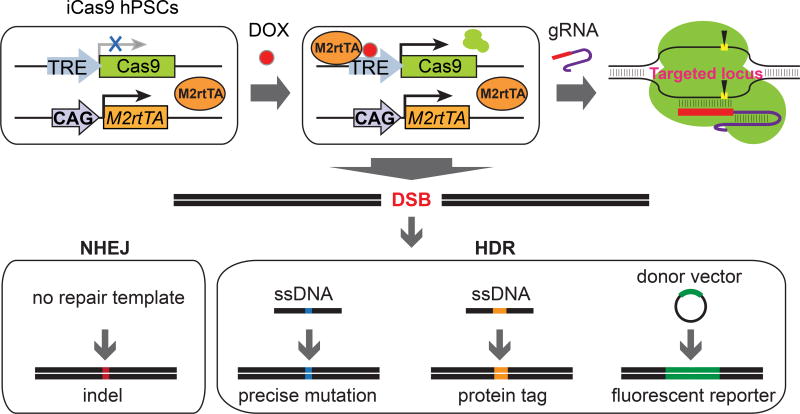
Figure 1. The iCRISPR platform for genome editing in hPSCs.
Doxycycline treatment induces Cas9 expression in iCas9 hPSCs. After transfection of gRNA, Cas9 is guided to the target locus via Watson-Crick base pairing and induces DNA DSBs. In the absence of a repair template, repair of the DSB by NHEJ often results in indels that can be used to knockout a target gene in hPSCs. Alternatively, in the presence of repair templates, either short ssDNA or long dsDNA donor vectors, HDR can be employed to incorporate exogenous sequences into the endogenous loci to introduce precise nucleotide mutations, protein tags or fluorescent reporters into the target loci.
3.1. hPSC Culture
hPSCs are cultured on an irradiated mouse embryonic fibroblast (iMEF) feeder layer in hPSC media.
iMEFs are plated 1 day before seeding hPSCs. Tissue culture dishes are coated with 0.1% gelatin for 30 minutes at 37°C prior to plating the iMEF feeder layer.
hPSCs cultures are generally passaged at 1:6 or 1:12 split ratios every 4–6 days. To passage hPSCs, colonies are disaggregated by treating with TrypLE Select enzyme for 5 minutes at 37°C.
Dissociated hPSCs are collected in hPSC media and spun down at 1,000 rpm for 5 minutes. After spinning hPSCs are resuspended in fresh hPSC media and seeded on iMEF coated plates. 5 µM ROCK inhibitor Y-27632 is added into the culture medium when passaging or thawing frozen cells.
Cell lines are frozen down in freezing media consisting of 40% FBS, 10% DMSO and 50% hPSC media.
3.2. Generation of knockin reporter hPSC lines using iCRISPR
The generation of knockin reporter hPSCs lines using the iCRISPR system involves first the induction of Cas9 expression through administration of doxycycline followed by the co-transfection of the gRNA and donor vector into the iCas9 cells. The transfected gRNA forms a complex with the Cas9 protein and directs it to the target locus, where it creates a DSB. HDR can then be employed to efficiently incorporate the exogenous reporter sequence into the target locus. After dissociating and re-plating, single-cell colonies are isolated, expanded and screened through PCR for correct reporter integration. Correctly targeted clonal lines are further validated and differentiated to assess faithful reporter activity (Fig. 2).
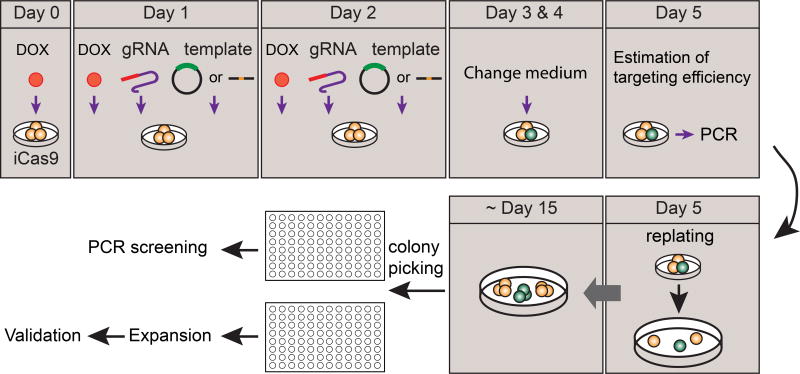
Figure 2. The workflow for iCRISPR-mediated knockin in hPSCs.
Doxycycline treatment 24 hours before the first transfection induces Cas9 expression in iCas9 hPSCs at Day 0. Co-transfection of gRNA and repair templates, either donor vectors or ssDNAs on Day 1 and Day 2 results in the incorporation of exogenous sequences into the endogenous loci. After PCR genotyping on Day 5, cells with the highest targeting efficiency will be re-plated as single-cells at a low density (~2,000 – 5,000 cells /100 mm dish) and allowed to grow for ~ 10 – 15 days. On Day 15, single-cell colonies will be picked, amplified, passaged into duplicated wells and subject to PCR screening. Correctly targeted clonal lines will be expanded and further validated.
3.2.1. Donor vector design and construction
Traditionally, the donor plasmid contains two homology arms flanking a promoterless reporter gene and a drug-resistance cassette driven by a constitutively active promoter. The two arms contain sequences that are homologous to the target locus. The constitutively active drug-resistance cassette enables enrichment of correctly targeted clones. However to avoid interference between the drug resistance cassette and neighboring genes, the drug-resistance cassette needs to be removed after identification of correctly targeted clones. Due to the high efficiency of the iCRISPR system, knockin reporter lines can be generated without the use of a drug selection cassette. This significantly reduces the time and effort required for establishment of knockin reporter hPSC lines.
Choose the target site for reporter integration in a way that ensures faithful reporter expression as well as minimizes the potential impact on endogenous gene expression and/or protein function. There are two general strategies for reporter gene integration, protein fusion (also called translational fusion) and promoter fusion (also called transcriptional fusion) (Fig. 3). For protein fusion, a reporter gene is integrated into the same reading frame of the target gene, transcribed as a single mRNA driven by the endogenous promoter and expressed as a single fusion protein. The protein fusion reporter is useful for monitoring protein subcellular localization, protein dynamics and protein-protein interactions of the target gene. However, the fused reporter may affect the folding, stability and function of the endogenous protein. Without previous knowledge about the functional domain and protein structure of the target gene, we recommend inserting the reporter gene either at the N-terminus immediately after the start codon or at the C-terminus immediately after the coding sequence. For promoter fusion, a reporter gene is also integrated into the same reading frame and transcribed together with the target gene as a single mRNA driven by the endogenous promoter. However, different from protein fusion reporter, promoter fusion reporter is preceded by either an IRES (internal ribosome entry site) or a 2A peptide sequence (Fig. 3). Consequently, the reporter and target genes will be translated into two separate peptides. Promoter fusion reporter reflects the endogenous gene expression pattern and usually has minimal effects on the endogenous gene expression or protein function. However, it is not suitable to monitor the subcellular localization, protein dynamics and protein-protein interactions of the target gene. Whether to make a protein fusion or a promoter fusion reporter depends on the intended application. For monitoring gene expression during hPSC differentiation, we are primarily concerned with dynamic changes in gene expression and less about the subcellular localization of the protein. Thus for this purpose we generally use the promoter fusion strategy and integrating the reporter gene at the C-terminus immediately after the coding sequence of the endogenous gene as this will have the least effect on endogenous protein function (Fig. 4A).
After deciding the integration locus, PCR amplify the two homologous arms (HA-L and HA-R) flanking the integration locus using high fidelity Herculase II Fusion DNA Polymerase (Note 1) and clone into a backbone vector. We generally use pBluescript, which contains multiple cloning sites and is relatively small, as the backbone vector.
While the optimal length of the homologous arms has not been thoroughly investigated, approximately 500 – 1,000 bp of homology arms on each side of the reporter gene are recommended (Note 2).
Insert the reporter, for example a fluorescent protein reporter, preceded by a 2A self-cleavage sequence (2A–reporter) between the two homology arms. Make sure the 2A–reporter sequence is fused in-frame to the last coding sequence of the target gene (Fig. 4B).
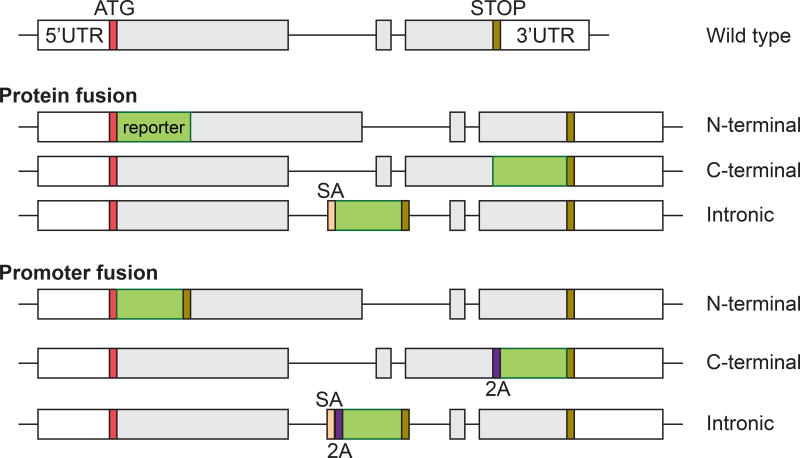
Figure 3. Schematics of protein and promoter fusion knockin strategies.
Here we illustrate typical targeting strategies for creating protein and promoter fusion knockin alleles. For a protein fusion reporter, the reporter gene (in green) is fused in-frame to the N-terminus of the endogenous gene immediately after the start codon (ATG, in red), to the C-terminus immediately after the last coding sequence or in the intronic region using a splicing acceptor sequence (SA, in orange). For a promoter fusion reporter, the reporter gene and the endogenous locus is linked through a 2A peptide (2A, in purple) and thus two separate peptides will be produced. The promoter fusion reporter can also be integrated into the N-terminus, C-terminus or intronic region of the endogenous gene. Boxes are exons, with open boxes indicating the untranslated region (UTR), filled grey boxes indicating the coding sequence (CDS); connecting lines are introns.
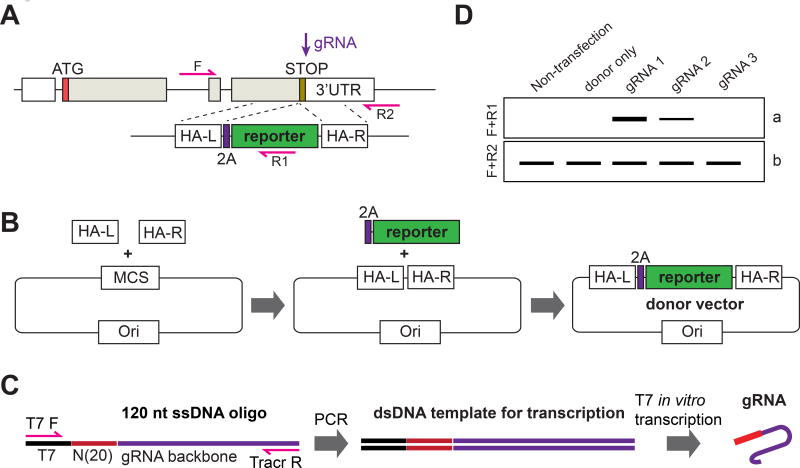
Figure 4. Generation of knockin reporter hPSC lines using iCRISPR.
A) Schematics of the targeting strategy. In the presence of the donor vector, HDR results in the replacement of stop codon with 2A–reporter sequence. The PCR primers (F + R1 and F+R2) used for genotyping are indicated with red arrows. (HA-L and HA-R indicate left and right homology arms). B) Schematics of the cloning strategy. The left and right homology arms (HA-L and HA-R) and the 2A–reporter gene are cloned sequentially into the backbone vector. (MCS: multiple cloning site). C) Schematics of the gRNA production. A 120 nt ssDNA containing a 20 nt T7 promoter sequence (T7, in black), a variable 20 nt gRNA recognition sequence ((N)20, in red) and a constant gRNA backbone sequence (in purple) is PCR amplified using T7 F and Tracr R universal primers (red arrows). The dsDNA PCR products will then be used as templates for in vitro transcription to produce the gRNAs. D) Estimation of targeting efficiency. The PCR primers (F+R1 and F+R2) are indicated in figure A. Comparison of the PCR product (band a) and (band b) allows an approximate estimation (a/(a+b) × 100%) of the targeting efficiency by each gRNA.
3.2.2. Design and production of gRNAs
We routinely use the CRISPR design tool developed by the Feng Zhang group at MIT (http://crispr.mit.edu/). The software not only identifies all possible CRISPR targets in an input DNA sequence but also uncovers potential off-target sites, thus predicting gRNAs with the highest targeting specificity. For each target locus, we recommend designing 3 gRNAs and selecting the gRNA that shows the highest targeting efficiency for establishment of clonal lines (estimation of the targeting efficiency is described in 3.2.3 #5).
We have found that it is faster and more cost effective to generate gRNAs using an oligonucleotide, than through cloning into the commonly used gRNA-expressing plasmids. First, a 120 nt ssDNA oligo, which includes a T7 promoter sequence, a variable 20 nt gRNA recognition sequence (N)20 (in red) and a constant gRNA backbone sequence can be directly synthesized and bought from commercial vendors. Next PCR amplify the oligonucleotide using the T7 F and Tracr R universal primers and use the PCR products as templates for in vitro transcription to produce the gRNAs (Fig. 4C).
Use the online CRISPR design tool (http://crispr.mit.edu/) to design 3 gRNAs that target sequences at the appropriate genomic site. It is highly recommended to choose gRNAs that do not recognize any sequence present in the donor vector to prevent undesired CRISPR/Cas9-mediated mutagenesis after correct reporter gene integration.
Order the 120 nt ssDNA oligos containing the desired 20 nt gRNA sequences directly from commercial vendors. Desalted purification is sufficient.
PCR amplify the 120 nt ssDNA oligos using the T7 F and Tracr R universal primers using Herculase II Fusion DNA Polymerase to produce dsDNA templates for T7 in vitro gRNA transcription.
-
Examine the PCR product through electrophoresis using 2.5% Agarose gel. Gel purify the desired dsDNA templates if non-specific PCR products are detected.
Primer Sequence T7 F TAATACGACTCACTATAGGG Tracr R AAAAGCACCGACTCGGTGCC PCR Reaction mix ( 50 µl)Component Amount ddH2O 35.5 µl 5X Herculase II reaction buffer 10 µl dNTP mix (25 mM) 0.5 µl T7 F (10 µM) 1.25 µl Tracr R (10 µM) 1.25 µl 120-nt ssDNA oligo (25 nM) 1 µl Herculase II fusion DNA polymerase 0.5 µl PCR cycling conditionsCycle number Denature Anneal Extend 1 94 °C , 2 min 2–31 94 °C, 20 sec 60 °C, 20 sec 72 °C, 1 min 32 72 °C, 2 min -
Use the MEGAshortscript T7 Transcription kit for gRNA synthesis.
In vitro transcription mix (20 µl)Component Amount T7 ATP 2 µl T7 CTP 2 µl T7 GTP 2 µl T7 UTP 2 µl T7 10X buffer 2 µl T7 enzyme mix 2 µl dsDNA template 8 µl Incubate for 4 hours to overnight at 37°C
Add 1 µl TURBO DNase and incubate for 15 minutes at 37°C to digest the dsDNA template.
Purify gRNA using the MEGAclear Transcription Clean-Up Kit following the manufacturer’s instructions, and elute gRNAs (typically ~ 50 – 100 µg) in 100 µl RNase-free water. When possible adjust concentration to 320 ng/µl (10 µM) and store at −80°C until use.
3.2.3. Co-transfection of gRNA and donor vector in iCas9 hPSC cells
Efficient co-transfection of gRNA and dsDNA donor vector into the iCas9 hPSC cells is very important for the generation of knockin reporter hPSC lines using iCRISPR system. We have tested several commonly used transfection reagents along with electroporation and find that the highest and most consistent co-transfection efficiency (~20%) is achieved using Lipofectamine 3000 (Note 3).
Day 0. Plate iMEFs on a gelatin-coated 24-well plate and treat 60% confluent iCas9 hPSC with hPSC medium containing 2 µg/ml doxycycline. This will allow optimal Cas9 expression at the time of transfection on Day 1.
-
Day 1. Wash the iCas9 cells with PBS w/o Ca2+ and Mg2+ and then dissociate hPSCs with TrypLE for 5 mins at 37°C. Once the cells are disaggregated, collect in 10 mL of hPSC media and spin down at 1000 rpm for 5 mins. Resuspend cells at ~ 0.25 – 1 × 106 cells/ml in hPSC medium with 5 µM ROCK inhibitor and 2 µg/ml doxycycline (Note 4). Re-plate 0.5 mL of cell suspension into each well of a 24-well plate. Prepare duplicated wells for each gRNA used, and wells for the non-transfection control and the donor-only control. One well of the duplicated wells will be used for estimation of the targeting efficiency and the other well will be used for re-plating.
1st transfection. For each gRNA, prepare separately:
Mix 1: 50 µl Opti-MEM + 1 µl gRNA (320 ng) + 2 µl P3000 + 2 µl donor plasmid (2.5 µg).
Mix 2: 50 µl Opti-MEM + 3 µl Lipo 3000 reagent.
Mix 1+2, incubate 5 min at RT and add 50 µl mixture dropwise into 0.5 ml hPSCs dilution in both duplicate wells.
For the non-transfection control, omit both gRNA and donor template.
For the donor-only control, add only donor template.
Day 2. Change hPSC medium with 2 µg/ml doxycycline and perform the 2nd transfection using the same procedure as the 1st transfection.
Day 3–4. Change hPSC medium daily.
-
Day 5. Estimation of targeting efficiency.
For lineage-specific genes that are only expressed in differentiated cell types, the targeting efficiency can be roughly estimated by PCR amplification of the targeted alleles. Collect genomic DNA from one of the duplicated wells using the DNeasy Blood & Tissue Kit. PCR amplify the targeted alleles using an external primer (F) and an internal primer (R1), and the non-targeted allele using two external primers (F and R2) (Fig. 4D). Although primers F+R2 may also amplify the targeted allele, in our experience the non-targeted allele is preferentially amplified due to its smaller size. Identify the gRNA with the highest targeting efficiency and use the corresponding well for expansion and screening of clonal lines.
PCR Reaction mix (50 µl)Component Amount ddH2O 35.5 µl 5X Herculase II reaction buffer 10 µl dNTP mix (25 mM) 0.5 µl Forward primer (10 µM) 1.25 µl Reverse primer (10 µM) 1.25 µl Genomic DNA (100 ng/µl) 1 µl Herculase II fusion DNA polymerase 0.5 µl PCR cycling conditionsCycle number Denature Anneal Extend 1 94 °C , 2 min 2–36 94 °C, 20 sec N* °C, 20 sec 72 °C, 30 sec 37 72 °C, 2 min *Annealing temperature of the primer pair, ideally between 55°C and 65°CFor genes that are expressed in undifferentiated hPSCs, the percentage of cells expressing the fluorescence reporter tag can be directly assessed through flow cytometric analysis.
3.2.4. Expansion and screening of clonal lines
-
Re-plating. hPSCs identified with the highest targeting efficiency in 3.2.3 are dissociated into single cells using TrypLE, re-suspended in hPSC medium with 5 µM ROCK inhibitor and re-plated at ~ 2,000 – 5,000 cells per 100 mm dish into dishes pre-seeded with iMEF feeders.
For genes that are expressed in undifferentiated hPSCs, enrich correctly targeted cells through Flow Activated Cell Sorting (FACS) and then re-plate them at a density of 10,000 cells per 100 mm dish.
Don’t change medium the next day.
Starting from 2 days after re-plating, change hPSC medium every day until colonies growing from single cells reach ~ 2 mm in diameter (~10 days).
-
Colony picking. Remove hPSC medium and add 10 ml PBS w/o Ca2+ and Mg2+ into the dish for a clearer visibility of the colonies. Mechanically dissociate the hPSC colonies into small clusters using 200 µl pipette tips, pick and plate single colonies in an uncoated 96-well plate containing 100 µl of hPSC medium with 5 µM ROCK inhibitor. Each clone is further disaggregated by pipetting up and down 5 times and re-plated in duplicated 96-well plates (50 µl each) pre-seeded with iMEFs and containing 100 µl of hPSC medium with 5 µM ROCK inhibitor. Alternatively, colonies could first be cultured in one 96-well plate and then split into two 96-well plates when confluent (in ~ 5 days).
Depending on the estimated targeting efficiency in 3.2.3, pick between 96 (targeting efficiency > 5%) to 384 clones (targeting efficiency < 5%). Media change and passaging using multichannel aspirator and dispenser pipettes greatly facilitates this step.
Change hPSC media for the 96-well plates daily. Use one of the duplicated 96-well plates for PCR screening in 3 days and the other plate for maintenance.
Genomic DNA extraction. Remove media from the wells, wash once with PBS w/o Ca2+ and Mg2+ and add 35 µl of genomic DNA lysis buffer. After 5 min at RT, transfer the cell lysates into a 96-well PCR plate. Seal the plate using a PCR film sticker and incubate at 55°C for 2 hours followed by 5 min at 95°C to inactivate Proteinase K. Keep samples at 4°C before the PCR screening step (Note 5).
PCR screening. PCR amplify using the same external primer (F) and internal primer (R1) used in 3.2.3. Run 25 µL of the PCR product in a 1% agarose gel stained with EtBr. PCR products of the right size suggest proper targeting in clonal lines.
Expansion of clonal lines. Amplify the desired clonal lines from the maintenance plate for further validation.
3.2.5. Validation of established clonal lines
After targeting, isolation of clonal lines and PCR genotyping, correctly targeted clonal lines are expanded and further validated. We recommend immunohistochemistry for expression of pluripotency markers (e.g. OCT4, SOX2 and NANOG), teratoma assay for a functional assessment of pluripotency, karyotype testing and differentiation into specific lineages to confirm faithful reporter activity. We highly recommend performing Sanger sequencing of both the targeted and the non-targeted allele at the target locus. We generally detect the expected sequence at the targeted allele. However, indel mutations are sometimes detected in the non-targeted allele, and clones with these undesired mutations should be discarded.
Due to mismatch tolerance of CRISPR gRNA paring (Hsu et al., 2013), there are concerns about the potential off-target mutagenic effects of the CRISPR/Cas9 system. The online CRISPR design tool (http://crispr.mit.edu/) can be used to identify the potential off-targets falling in coding sequences for each gRNA and we typically select 10 top candidates for further analysis. PCR amplify the genomic region (~500 bp) flanking the CRISPR off-target site using Herculase II Fusion DNA Polymerase and analyze by Sanger sequencing using a primer that binds within the PCR product. It is worth noting that our analysis so far hasn’t revealed any off-target mutations at sites without perfect complementarity to the CRISPR/Cas9 target sequence. In agreement with our findings, a recent high-coverage whole-genome sequencing study failed to detect significant incidence of off-target mutations in CRISPR-targeted hPSC lines (Veres et al., 2014).
3.3. Generation of knockin epitope tagged hPSCs using iCRISPR
The procedure to generate knockin epitope tagged hPSCs is similar to the procedure used to create reporter tags (Section 3.2). Expression of Cas9 is induced in iCas9 cells following administration of doxycycline. A gRNA for the target site and the exogenous sequence are delivered into the cells. Site-specific DSBs produced by Cas9 promote HDR and integration of the epitope tag into the genome. After two transfections the cells are re-plated at clonal densities. Clonal lines are then isolated, expanded and genotyped using PCR.
One key difference from Section 3.2 is that due to the smaller size of the epitope sequence, a ssDNA can be used to provide the HDR template (the epitope tag flanked by the homology arms). In addition a different transfection reagent, Lipofectamine RNAiMax is used to deliver the ssDNA and gRNA into the cells. Finally clonal lines identified by PCR to have correct integration, are further validated using Sanger sequencing, IP and western blot.
3.3.1. Design of CRISPR gRNAs and donor ssDNA
For the generation of tagged proteins the location of the tag in reference to the protein is very important. The tag should be located so as to have minimal effect on tertiary structure and biological activity of the protein. At the same time the tag should be readily accessible on the surface of the natively folded protein. In deciding where to insert the tag it is best to follow previous studies that have already generated the fusion protein for functional studies. In the absence of a previous study, the safest strategy would be generate lines that have the tag epitope attached to the N-terminal of the target protein, as well as lines that have the tag epitope attached to the C-terminal of the target protein (Fig. 5A). After these lines have been generated one can compare them to see if the tag influences the stability or biological function of the protein. This could be done using western blot, co-IP with known interacting proteins, and for DNA-binding proteins, ChIP-qPCR.
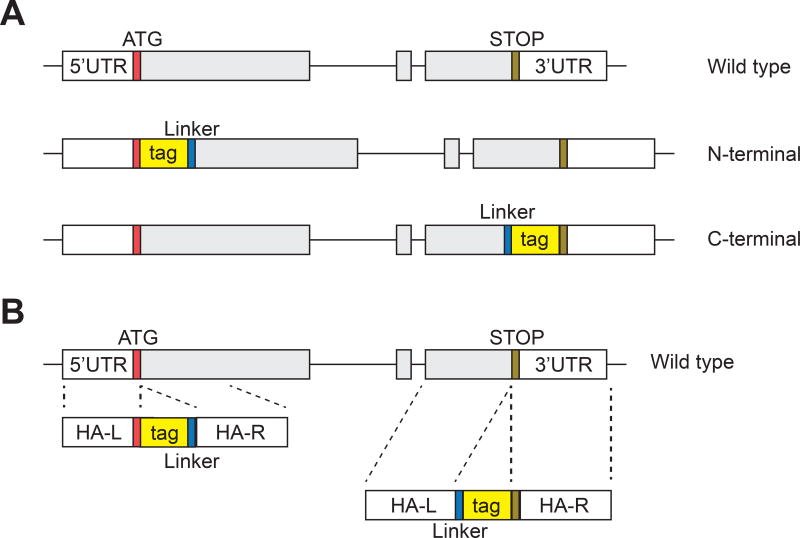
Figure 5. Generation of epitope tag knockin hPSC lines using iCRISPR.
A) Knockin epitope tags can be targeted to the N-terminal or C-terminal of a protein. At the N-terminal the epitope tag is inserted right after the start codon and separated from the endogenous sequence by a linker sequence. At the C-terminal the epitope tag is inserted immediately before the stop codon with a linker sequence separating the epitope tag from the endogenous sequence. B) Schema of the ssDNA donors used to generate knockin epitope tags.
When targeting the N-terminal of the protein, 3–4 gRNAs are chosen to cut near the start codon, so that after integration the start codon will immediately precede the epitope tag. When targeting the C-terminal of a protein, 3–4 gRNAs are designed to cut near the stop codon of the protein, so that after integration the epitope tag will be immediately before the stop codon (Fig. 5A). Please refer to section 3.2.2 for detailed instructions on the design and production of CRISPR gRNAs.
3.3.2. Design of donor ssDNA
The donor template consists of 3 components: the left and right homology arms, the epitope tag sequence and a short linker sequence. The smaller size of the epitope tag compared to a reporter transgene, allows smaller homology arms (less than 100 nt) to be used for the targeting. Thus the overall size of the donor template is typically between 150 and 300 nt, and a ssDNA donor, instead of a plasmid, can be used for the targeting experiment.
Each of the homology arms (left and right) is around 65 nt in length (Note 6).
For targeting the N-terminus, the left homology arm should stop after the start codon of the protein, and the right homology arm should include the DNA sequence immediately after the start codon (Fig. 5B). The sequence of the epitope tag is inserted between the homology arms (Note 7), followed by a short linker sequence (e.g. 12 nt long). The linker sequence separates the epitope tag from the endogenous protein. This prevents the epitope tag from interfering with proper folding of the endogenous protein (Borjigin and Nathans, 1994), and increases the accessibility of the epitope to antibodies (Grote et al., 1995). A linker sequence we have used is: GGAGGACTCGAC, which produces a 4 amino acid sequence of Gly-Gly-Leu-Asp.
If targeting the C-terminus, the left arm should contain the sequence up to, but not including, the stop codon. This is followed by the linker sequence and then the epitope tag. Lastly the right homology arm should begin with the stop codon (Fig. 5B).
The final ssDNA oligo, containing the two homology arms, a linker sequence and the epitope tag, can be ordered directly from commercial vendors. Desalted purification is sufficient.
3.3.3. Co-transfection of donor ssDNA and CRISPR gRNA
The procedure for passaging the cells for co-transfection is the same as described in 3.2.3. However, the transfection mixture is different. Perform co-transfection of donor ssDNA and CRISPR gRNA as follows:
Mix 1: 50 µl Opti-MEM + 1 µl gRNA (320 ng) + 5 µl ssDNA (1.5 µg).
Mix 2: 50 µl Opti-MEM + 3 µl Lipofectamine RNAiMAX.
Mix 1+2, incubate 5 min at RT and add 50 µl mixture dropwise into 0.5 ml dissociated hPSCs in one well of a 24-well plate.
The same procedure presented in Section 3.2.3 #5 can be used to evaluate the targeting efficiency for the epitope tag.
3.3.4. Expansion and PCR based-screening of clonal lines
The procedure for isolating clonal lines, PCR screening and expansion of clonal lines is the same as described in Section 3.2.4.
3.3.5. Sequencing verification of targeted clones
We highly recommend examining both the targeted and non-targeted allele at the target locus by Sanger sequencing. This enables us to verify that the epitope tag sequence is intact and properly integrated in-frame with the protein sequence. We also verify that the non-targeted allele does not contain any unwanted mutations.
Collect genomic DNA of correctly targeted lines using the DNeasy Blood & Tissue kit.
-
PCR amplify the targeted and non-targeted alleles using a pair of external primers.
PCR Reaction mix (50 µl)Component Amount ddH2O 35.5 µl 5X Herculase II reaction buffer 10 µl dNTP mix (25 mM) 0.5 µl Forward primer (10 µM) 1.25 µl Reverse primer (10 µM) 1.25 µl Genomic DNA (100 ng/µl) 1 µl Herculase II fusion DNA polymerase 0.5 µl PCR cycling conditionsCycle number Denature Anneal Extend 1 94 °C , 2 min 2–36 94 °C, 20 sec N* °C, 20 sec 72 °C, 30 sec 37 72 °C, 2 min *Annealing temperature of the primer pair, ideally between 55°C and 65°C Run 25 µl of the PCR reaction on a 1% agarose gel stained with EtBr.
Visualize the gel using a UV lamp. For correctly targeted clones there should be two bands, representing the targeted allele and the non-targeted allele. The targeted allele should be larger than the non-targeted allele by the size of the epitope tag (generally less than 100 bp).
Cut out the individual bands and purify DNA using the Purelink Quick Gel Extraction kit.
Use one of the PCR primers for Sanger sequencing, or use a new sequencing primer that is within the PCR product (Note 8).
3.3.6 Validation of targeted clones using immunoprecipitation and western blot
The final, and most important test, to validate the epitope tagged lines is to perform IP for the tag and then verify that the target protein is also pulled down. This would involve first an IP using an antibody against the epitope tag followed by a western blot with the antibody to the protein of interest. If it is challenging to detect the target protein due to lack of quality antibodies, one may perform the western blot against the tag epitope to show that the epitope tagged protein is the same size as the target protein. Below we provide a protocol routinely used in our laboratory for detection of 3xFLAG tagged proteins. We strongly encourage users to optimize the protocol for their protein of interest, as it may require different procedures for cell lysis, IP and western blot.
Expand the clonal lines into a 100 mm dish.
Use the Pierce Crosslink Magnetic IP/Co-IP kit for lysis and IP and follow the manufacturer’s protocol. Lyse one 100 mm dish per IP with 600 µL of the provided lysis IP buffer. Incubate the cell lysate with the FLAG antibody for 16 hours at 4°C. And elute with 100 µL of the provided elution buffer.
For the western blot 30 µL total volume is run: 19.5 µL of IP sample, 7.5 µL LDS sample buffer and 3 µL of reducing agent. The samples should be denatured for 10 minutes at 70°C prior to loading.
Samples are run on NuPAGE Novex 3–8% Tris-Acetate Protein Gels in NuPAGE Tris-Acetate SDS Running Buffer (20X) for 1 hour at 120 V.
Transfer is performed overnight at 30 V onto a nitrocellulose membrane.
Wash the membrane in TBST and then incubate in blocking buffer (5% non-fat milk in TBST) for 30 minutes at room temperature.
Incubate the membrane with the FLAG antibody at a 1:2,000 dilution overnight at 4°C.
Wash the membrane 3 times for 5 minutes each with TBST, and then incubate with HRP conjugated anti-mouse antibody (1:5,000) for 1 hour at room temperature.
Finally develop the membrane using ECL reagent. Prior to exposure, excess liquid should be removed without drying the membrane. Transfer the membrane to a clean sealed bag. Expose onto an autoradiography film in a hypercassette.
4. Notes
Undesired mutation in homology arms induced during PCR amplification may decrease the targeting efficiency. We recommend using the high fidelity Herculase II Fusion DNA Polymerase for PCR amplification of homology arms and also later for PCR amplification of the gRNA in vitro transcription template. One may substitute Herculase with other commercially available high fidelity DNA polymerases.
Traditional targeting procedures that do not utilize site-specific nucleases require homology arms of at least 5 – 12 kb for the insertion of reporter and selection cassettes and the targeting efficiency can be further increased by using substantially longer homology regions (up to 100 kb) (Nieminen et al., 2010). In most traditional targeting experiments, the donor plasmid typically contains a long (~ 10 kb) and a short homology arm (~ 2 – 4 kb) for efficient homologous recombination and also for convenient identification of correctly targeted alleles by PCR genotyping at the shorter homology arm side. With the use of engineered DNA endonucleases, repair templates with much shorter homology arms (~ 500 – 1,000 bp on each side) have been used. We have found that approximately 500 bp combined length of the homology arms (~ 250 bp on each side) is necessary to support efficient CRISPR-mediated targeting of a 750 bp exogenous sequence into an endogenous locus.
In addition to Lipofectamine 3000, a number of other transfection methods have also been shown to work efficiently in hPSCs (Byrne et al., 2015; Merkert et al., 2014).
Based on our experience with HUES8 and MEL-1 hESCs, we recommend testing several cell densities for the co-transfection of gRNA and donor vector, for example plating 100, 250 and 500K cells per well in a 24-well plate and choosing the best condition for the establishment of clonal lines. After transfection, we regularly use a replating cell density of 2,000–5,000 cells per 100 mm dish. These conditions may need to be adjusted for other hPSC lines.
This simplified protocol enables the extraction of genomic DNA for PCR screening without the use of phenol/chloroform extraction and purification. Combined with the use of multichannel pipettes, one can quickly screen a large number of clones. Based on our experience it is feasible for a single person to process up to 384 samples (four 96-well plates) at a time. This procedure works well as long as the primers are pre-tested before the targeting experiment.
We have used homology arms that are around 65 nt in length. The targeting efficiency may be increased by using longer homology arms, but this has not been systematically tested.
Thus far we have tested attaching a 3xFLAG epitope tag to the protein of interest. However this strategy can be used for attaching a variety of other epitope tags such as poly-Arg, poly-His, Strep-tag, c-myc-tag, HAT-tag, and His-tag. As long as the sequence of the tag remains under 100 bp the same procedure as presented above can be used. For larger tags the size of the homology arms and the delivery of the exogenous sequence (as a ssDNA or as a donor plasmid) may need to be adjusted. The specific protein tag to use will depend on the protein of interest as well as the intended downstream analysis.
If a nonsynonymous mutation is detected in the coding sequence of the protein in either the targeted or non-targeted allele we would advise to discard that particular clone. In some cases a mutation may occur in a non-coding regulatory region (e.g. promoter), in which case the clone may still be used if the mutation does not alter the expression of the affected allele (as determined by qRT-PCR for example).
5. Future Applications
Here we present the protocols to integrate epitope and reporter tags within the genome of hPSCs using the iCRISPR genome editing platform. The same knockin strategy could be used for additional purposes. For example, targeting a fluorescent or antibiotic tag into the coding region of a gene could be used to generate genetic knockouts. Specific deletions could be engineered to mimic a genetic disease or to dissect the contribution of particular protein domains for protein function. By inserting particular sequences one may even precisely modulate the DNA-binding, protein interaction or other specific activities of endogenous proteins. Furthermore, the gene targeting strategies presented here are also relevant to labs that use engineered nucleases other than CRISPR/Cas or employ different strategies to deliver these nucleases into hPSCs. Finally we provide robust protocols to achieve relatively high rates of delivery for ssDNA, dsDNA plasmids and gRNAs. These transfection procedures may have wider applicability such as potential uses in overexpression and lineage tracing studies.
Acknowledgments
Our work related to this publication was funded in part by NIH (R01DK096239) and NYSTEM (C029156). Z.Z. was supported by the New York State Stem Cell Science (NYSTEM) fellowship from the Center for Stem Cell Biology (CSCB) of the Sloan Kettering Institute. N.V. was supported by the Howard Hughes Medical Institute (HHMI) Medical Research.
References
- Borjigin J, Nathans J. Insertional mutagenesis as a probe of rhodopsin’s topography, stability, and activity. J Biol Chem. 1994;269:14715–14722. [PubMed] [Google Scholar]
- Byrne SM, Ortiz L, Mali P, Aach J, Church GM. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015;43:e21. doi: 10.1093/nar/gku1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RP, Costa M, Grandela C, Holland AM, Hatzistavrou T, Micallef SJ, Li X, Goulburn AL, Azzola L, Elefanty AG, et al. A protocol for removal of antibiotic resistance cassettes from human embryonic stem cells genetically modified by homologous recombination or transgenesis. Nat Protoc. 2008;3:1550–1558. doi: 10.1038/nprot.2008.146. [DOI] [PubMed] [Google Scholar]
- Eiges R, Schuldiner M, Drukker M, Yanuka O, Itskovitz-Eldor J, Benvenisty N. Establishment of human embryonic stem cell-transfected clones carrying a marker for undifferentiated cells. Curr Biol. 2001;11:514–518. doi: 10.1016/s0960-9822(01)00144-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR Platform for Rapid, Multiplexable, and Inducible Genome Editing in Human Pluripotent Stem Cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E, Hao JC, Bennett MK, Kelly RB. A targeting signal in VAMP regulating transport to synaptic vesicles. Cell. 1995;81:581–589. doi: 10.1016/0092-8674(95)90079-9. [DOI] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkert S, Wunderlich S, Bednarski C, Beier J, Haase A, Dreyer AK, Schwanke K, Meyer J, Gohring G, Cathomen T, et al. Efficient designer nuclease-based homologous recombination enables direct PCR screening for footprintless targeted human pluripotent stem cells. Stem Cell Reports. 2014;2:107–118. doi: 10.1016/j.stemcr.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Neuhausser WM, Santos D, Valen E, Gagnon JA, Maas K, Sandoe J, Schier AF, Eggan K. Efficient CRISPR-Cas9-Mediated Generation of Knockin Human Pluripotent Stem Cells Lacking Undesired Mutations at the Targeted Locus. Cell Rep. 2015;11:875–883. doi: 10.1016/j.celrep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen M, Tuuri T, Savilahti H. Genetic recombination pathways and their application for genome modification of human embryonic stem cells. Exp Cell Res. 2010;316:2578–2586. doi: 10.1016/j.yexcr.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabar V, Studer L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat Rev Genet. 2014;15:82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Gonzalez F, Huangfu D. The iCRISPR Platform for Rapid Genome Editing in Human Pluripotent Stem Cells. Methods Enzymol. 2014;546:215–250. doi: 10.1016/B978-0-12-801185-0.00011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Huangfu D. Human pluripotent stem cells: an emerging model in developmental biology. Development. 2013;140:705–717. doi: 10.1242/dev.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Verma N, Gonzalez F, Shi ZD, Huangfu D. A CRISPR/Cas-Mediated Selection-free Knockin Strategy in Human Embryonic Stem Cells. Stem Cell Reports. 2015;4:1103–1111. doi: 10.1016/j.stemcr.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]