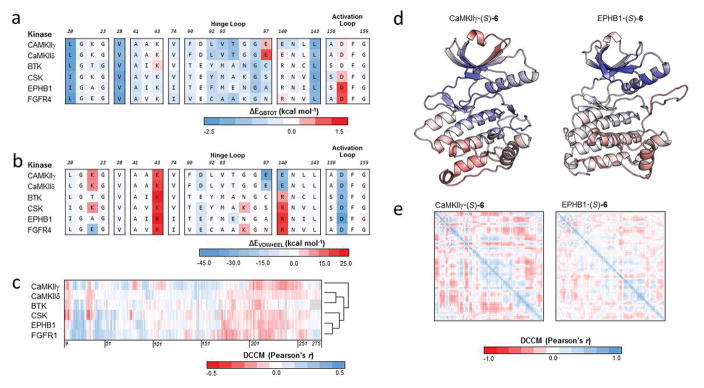
Figure 7.
(a) Total decomposition energy (ΔEGBTOT) of key residues of binding for (S)-6 against 6 kinase targets. Each row is a kinase target while each column is an aligned residue. (b) Summation of the van der Waals (ΔEVDW) and electrostatic (ΔEELE) terms from the total decomposition energy of key residues of binding for (S)-6 against the same 6 kinase targets. (c) Dynamic cross-correlation matrix (DCCM) cross section of (S)-6 against the protein kinase domain of 6 kinase targets. Gaps in the multiple sequence alignment are shown in gray. Cross sections were converted into vectors and hierarchically clustered using Euclidean distance and average linkage. (d) The correlation of motion of 6S bound to CaMKIIγ and EPHB1 were mapped onto the structures of the protein kinase domains. Correlation coefficients were colored according to the scale in panel c. (e) Per-residue DCCM of the protein kinase domain for (S)-6 bound to CaMKIIγ and EPHB1.