Abstract
Celecoxib is known to alter the preferred position of SULT2A1-catalyzed sulfonation of 17β-estradiol (17β-E2) and other estrogens from the 3- to the 17-position. Understanding the effects of celecoxib on estrogen sulfonation is of interest in the context of the investigational use of celecoxib to treat breast cancer. This study examined the effects on celecoxib on cytosolic sulfotransferases in human and rat liver and on SULT enzymes known to be expressed in liver. Celecoxib’s effects on the sulfonation of several steroids catalyzed by human liver cytosol were similar but not identical to those observed previously for SULT2A1. Celecoxib was shown to inhibit recombinant SULT1A1-catalyzed sulfonation of 10 nM estrone and 4 μM p-nitrophenol with IC50 values of 2.6 and 2.1 μM, respectively, but did not inhibit SULT1E1-catalyzed estrone sulfonation. In human liver cytosol, the combined effect of celecoxib and known SULT1A1 and 1E1 inhibitors, quercetin and triclosan, resulted in inhibition of 17β-E2-3-sulfonation such that the 17-sulfate became the major metabolite: this is of interest because the 17-sulfate is not readily hydrolyzed by steroid sulfatase to 17β-E2. Investigation of hepatic cytosolic steroid sulfonation in rat revealed that celecoxib did not stimulate 17β-E2 17-sulfonation in male or female rat liver as it does with human SULT2A1 and human liver cytosol, demonstrating that rat is not a useful model of this effect. In silico studies suggested that the presence of the bulky tryptophan residue in the substrate-binding site of the rat SULT2A homolog instead of glycine as in human SULT2A1 may explain this species difference.
Keywords: Sulfotransferases, Estrogen, Steroids, Celecoxib, Quercetin, Triclosan, Cancer
1. INTRODUCTION
Sulfotransferases (SULTs) are a complex enzyme family performing important functions, including molecular recognition, detoxification, hormone regulation, drug processing, and modulation of receptor binding [1, 2]. SULTs have also been implicated in a number of disease states including cancer metastasis and hormone-dependent breast tumor growth [3, 4]. As estrogen sulfates do not interact with estrogen receptors, SULT activity reduces estrogen-stimulated cancer cell growth [5]. SULTs are present in many tissues including the liver, kidney, brain, mammary glands, adrenal glands, intestine and platelets. In humans, at least 12 cytosolic SULT isoforms have been identified and divided into several gene families based on their amino acid sequences [6]. The most studied SULT forms are SULT1A1, 1A3, 1B1, 1E1 and 2A1. Of these, SULT1E1, 1A1 and 2A1 are expressed in human liver and can metabolize 17β-estradiol (17β-E2; estra-1,3,5(10)-triene-3,17 β-diol) [7, 8].
Celecoxib, a cyclooxygenase-2 (COX-2) inhibitor, is a nonsteroidal anti-inflammatory drug (NSAID) that is used for the relief of pain, fever, swelling, and tenderness caused by osteoarthritis and rheumatoid arthritis. Apart from acting as an anti-inflammatory drug, celecoxib has also shown effectiveness in prevention and treatment of proliferative diseases. For example, celecoxib reduced polyp size and progression to adenomas in patients with familial adenomatous polyposis [9]. Inhibition of COX-2 is one mechanism by which celecoxib is chemopreventive in patients with familial adenomatous polyposis, however other mechanisms may contribute to celecoxib’s efficacy. Celecoxib induced apoptosis in breast cancer cells in an in vivo model and inhibited growth of breast epithelial cells [10, 11]. Combinations of the ether analog of RRR-alpha-tocopherol (vitamin E) with celecoxib synergistically inhibited tumor volume compared to individual treatments with either drug in nude mice [12]. Studies in rats have found that a combination of the aromatase inhibitor exemestane with celecoxib was better at slowing the growth of breast cancer than either drug used alone [13]. A human clinical trial reported anti-tumor effects of celecoxib treatment in breast cancer patients [14]. It was speculated that COX-2 inhibition may contribute to the prevention and/or treatment of breast cancer by celecoxib [13–15] however, the mechanisms underlying its antitumor activity are not fully understood.
The ability of celecoxib to switch the dominant product of 17β-E2 sulfonation from 3-sulfate to 17-sulfate with human recombinant SULT2A1 as well as in human liver cytosol [8] suggested that 17β-E2 levels in breast tissue would be reduced by celecoxib, as the 17-sulfate was resistant to sulfatase hydrolysis [16, 17]. This suggests another mechanism by which celecoxib could be beneficial in treating estrogen receptor positive breast cancer.
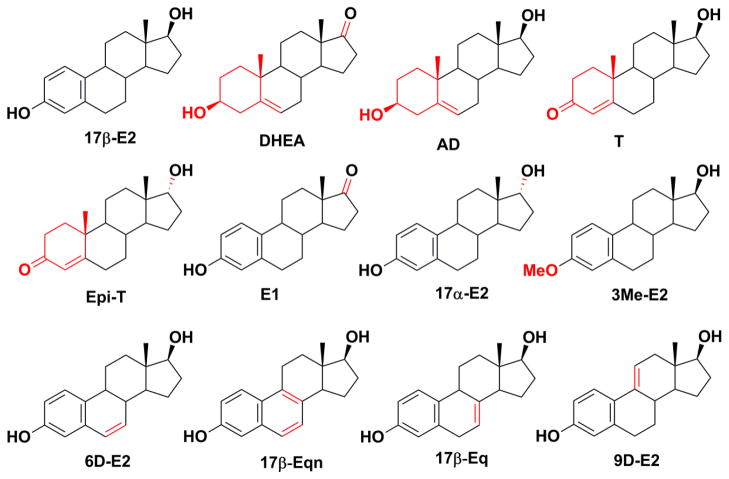
The effects of celecoxib on the position and rate of sulfonation of several steroids, shown in Figure 1, have been examined with human recombinant SULT2A1 [18]. They are dehydroepiandrosterone (DHEA; (3β)-3-hydroxyandrost-5-en-17-one), androstenediol (AD; (3β,17β)-androst-5-en-3,17-diol), epitestosterone (Epi-T; (17α)-17-hydroxyandrost-4-en-3-one), testosterone (T; (17β)-17-hydroxyandrost-4-en-3-one), 17α-estradiol (17α-E2; (17α)-estra-1,3,5(10)-triene-3,17-diol), estrone (E1; 3-hydroxyestra-1(10),2,4-trien-17-one), 3-methyl ether of 17β-E2 (3Me-E2), 6-dehydroestradiol (6D-E2; estra-1,3,5(10),6-tetraene-3,17-diol), 17β-dihydroequilenin (17β-Eqn; 1,3,5(10)6,8-estrapentaen-3,17 β-diol),17β-dihydroequilin (17β-Eq; 1,3,5(10)7-estratetraen-3, 17β-diol) and 9-dehydroestradiol (9D-E2; (17β)-estra-1,3,5(10),9(11)-tetraene-3,17-diol). With 17β-E2, 6D-E2, 9D-E2 and the equine estrogens 17β-Eqn and 17β-Eq that are found in hormone replacement therapy preparations such as premarin [19], celecoxib switched the preferred position of sulfonation by SULT2A1 from the 3-OH to the 17β-OH. The modulation of SULT2A1 activity observed was proposed to be due to a conformational change of the SULT2A1 dimer upon celecoxib binding to the PAPS (3′-phosphoadenosine-5′-phosphosulfate) binding site in one of the constituent monomers such that the 17β-estrogens bound with the 17β-OH in a favorable position for sulfonation. The stimulation of 17-sulfonation occurred regardless of the order of addition of celecoxib or PAPS to assays. The effect of celecoxib in human liver cytosol, which is an important site of steroid sulfonation, is of interest for understanding the likely in vivo effects of celecoxib on metabolism of physiologically important estrogens and the equine estrogens commonly used in hormone replacement therapy.
Figure 1.
Chemical structures of steroids tested for the effect of celecoxib on their sulfonation by human liver cytosol. The structural differences with 17β-E2 are highlighted in red.
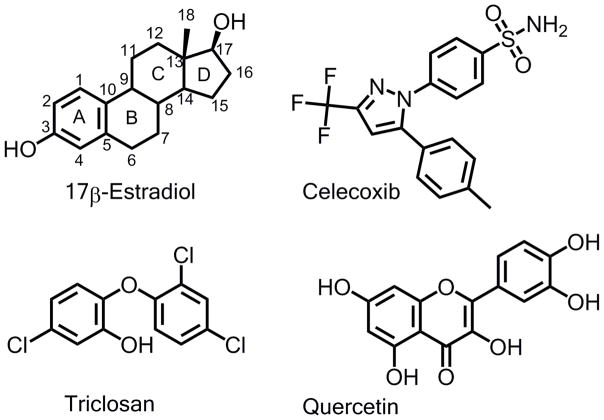
It was previously shown that the ratio of 17β-E2–17-sulfate/17β-E2–3-sulfate was higher for human SULT2A1 than for human liver cytosol at a given concentration of celecoxib, and this was thought to be due to the contributions of SULT1A1 and SULT1E1 to 17β-E2–3-sulfate formation in liver [8]. It is known that several xenobiotics including dietary and environmental chemicals are better inhibitors of SULT1A1 and SULT1E1 than of SULT2A1 [20, 21]. We hypothesized that the presence of specific phenol SULT inhibitors as well as celecoxib in incubations with human liver cytosol would result in increases in the ratio of 17-sulfate/3-sulfate. In this study, we investigated the sulfonation of 17β-E2 in the presence of both celecoxib and the phenol SULT inhibitors, triclosan or quercetin. Quercetin was reported as a potent inhibitor of SULT1A1 [20–23] while triclosan inhibited SULT1E1 and 1A1 [20, 21, 24, 25]. The structures of inhibitors are shown in Figure 2.
Figure 2.
Structure of 17β-E2 with carbon atoms numbered and rings named according to steroid nomenclature. Structure of celecoxib and the SULT inhibitors triclosan and quercetin are also shown.
The switching of the major product of 17β-E2 sulfonation by SULT2A1 in the presence of celecoxib prompted studies in the widely used model animal, the rat, to investigate if this effect is observed in other species. Considering that rat liver is sexually dimorphic for the expression of sulfotransferases [26, 27], both female and male rat liver cytosolic preparations were tested separately for their activity towards steroid substrates in the presence and absence of celecoxib. For the species comparison, we selected substrates that showed isomer-selective differences of sulfonation catalyzed by SULT2A1 [18]. These were T and Epi-T, 17β-E2 and 17α-E2. DHEA and AD were also selected because both are converted to 3β-sulfate by SULT2A1, even though AD also has a 17-OH group.
The goals of this study were to investigate: (1) the effect of celecoxib on human hepatic cytosolic sulfonation of non-aromatic steroids and estradiol analogs (2) the combined effect of celecoxib and selected phenol sulfotransferase inhibitors on 17β-E2 sulfonation by human liver cytosol and (3) how celecoxib affected rat liver cytosolic sulfotransferase activities toward steroids.
2. MATERIALS AND METHODS
2.1. Reagents
The structures of the steroids and inhibitors used in this study are as shown in Figures 1 and 2. p-Nitro[U-14C]phenol, 118 μCi/μmol (98.4% pure) was obtained from Amersham BioSciences UK Ltd. The 14C-p-nitrophenol (pNP) was diluted with unlabeled pNP to a specific radioactivity of 20.1 μCi/μmol for use. Quercetin (spectrophotometric grade) was obtained from Sigma (St. Louis, MO). Celecoxib was extracted from capsules and isolated as described previously and had no detectable impurities [8]. The [35S]-PAPS (1.86 Ci/mmol) was from PerkinElmer Life Sciences, Inc., (Boston, MA). Unlabeled steroids were obtained from Steraloids (Newport, RI). Unlabeled PAPS was purchased from Dr. Sanford S. Singer (University of Dayton, OH) and from Sigma-Aldrich (St. Louis, MO) and was purified as described previously [28]. The 17β-E2–3-sulfate standard was obtained from Sigma-Aldrich (St. Louis, MO). The other steroid sulfate standards were from Steraloids (Newport, RI).
2.2. Cytosolic preparations
The human liver samples from organ donors were kindly supplied by Dr. F. Peter Guengerich (Vanderbilt University, Nashville, TN) under an exempt Institutional Review Board protocol. Liver cytosolic preparations were made by standard differential centrifugation methods as described previously [29]. A pool of liver cytosol from three individuals (two males, ages 20 and 45 years and one female, 27 years of age) was used, with the same protein amount from each. Cytosol fractions from three individual 16-week old female and male Sprague Dawley rat livers were used for in vitro rat experiments. The rats were untreated control rats from other studies as approved by the University of Florida Institutional Animal Care and Use Committee.
2.3. Sulfotransferase assays
Human recombinant SULT1A1*1, 1A3, 1B1, 1E1 and 2A1 were prepared and isolated as described previously [8, 18]. Assay conditions with all substrates were optimized such that the rate of reaction was linear with protein and time, and was saturating for PAPS. In all studies, duplicate determinations were made for each condition.
The assay conditions with the steroid substrates were as described previously for SULT2A1 [18] except that 0.01–0.15 mg pooled human liver cytosol was used for 0.4 μM DHEA, AD, Epi-T, T, 17β-E2, 17α-E2, 3Me-E2, 6D-E2, 9D-E2, 17β-Eqn and 17β-Eq and 0.005–0.1 mg cytosol from rat livers was used for in vitro rat experiments. The reaction products of steroid sulfonation were analyzed by HPLC and LC-MS/MS as described previously [18].
To study the sulfonation of 10 nM E1, 0.012 mg SULT1A1*1 or 0.004 mg SULT1E1 was used [18]. Sulfotransferase activity with pNP was measured with 0.007 mg SULT1A1*1 or 0.03 mg pooled human liver cytosol, 100 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 4 μM of 14C-pNP and 20 μM PAPS (added last to start the reaction), in a total volume of 250 μL [24]. It has been reported that although pNP can be sulfonated by several SULTs, it is selective for SULT1A1 at 4 μM [30]. Stock solutions of celecoxib, quercetin and triclosan were prepared in DMSO, such that the total DMSO concentration in assays did not exceed 0.5% (v/v).
2.4. Homology modeling and ligand-docking
The most abundant form of hydroxysteroid sulfotransferase in female rat liver [26], ST-60 (NCBI accession number NP_001020302.1), was used for homology modeling with SULT2A1 crystal structure (PDB ID 1J99) and the 3D-model was treated and used for the ligand docking experiments as described previously [18]. Briefly, hydrogen atoms and Amber7 FF99 charges were added to the model predicted with the help of SWISS-MODEL [31, 32] and a staged minimization in Amber7 FF99 force field was performed. Analysis by SAVES (The Structure Analysis and Verification Server run by Molecular Biology Institute at the University of California, Los Angeles) showed that the 3D structures passed Errat, Verify3D and Procheck and had no residues with disallowed Ramachandran conformations.
Flexidock utility in Sybyl-X 2.0 suite was used for the ligand docking experiments with 17β-E2 and celecoxib as described previously [18]. Our objective was to determine the likely effect of celecoxib binding in the substrate binding site of human and rat orthologs of hydroxysteroid sulfotransferase on steroid sulfonation. Figures of the docking results were generated by Chimera 1.6.2. [33, 34].
2.5. Data analysis
Results for the pool of human liver cytosol and for the expressed enzymes are from the mean of duplicate determinations. IC50 values were obtained from non-linear regression analysis of the relationship between the percentage of control activity and log inhibitor concentration using GraphPad Prism 6 software (Menlo Park, CA). For rat experiments duplicate measurements were made with each of the three female and male animals and the data presented is the mean ± SD of all six measurements with each substrate.
3. RESULTS
3.1. Effects of celecoxib on the sulfonation of non-aromatic steroids
Among the steroids tested, Epi-T, DHEA and AD were the most sulfonated substrates by human liver cytosol at a concentration of 0.4 μM with control activities of 70.6, 50.2 and 40.9 pmoles/min/mg protein, respectively (Figure 3A and 3B). Epi-T was sulfonated with about 17.5 times higher activity than its isomer at C-17 position, T (activity was 4.0 pmoles/min/mg protein). In the presence of increasing concentrations of celecoxib, the sulfonation of the non-aromatic steroids was inhibited with IC50 values of 53.5, 58.5, 67.4 and 55.5 μM for DHEA, AD, Epi-T and T, respectively. As in the case of recombinant SULT2A1 [18], human liver cytosol also catalyzed the formation of just one of the two possible sulfates of AD.
Figure 3.
Effect of celecoxib on the sulfonation of DHEA, AD, Epi-T, T 17α-E2 and E1. (A) DHEA–sulfate (●) and AD–sulfate (○). (B) Epi-T–sulfate (●) and T–sulfate (○). All the substrates at 0.4 μM concentration were incubated with the amount of pooled human liver cytosol required to convert 20–40 % of them to their respective sulfate products. The analysis of the products was conducted as described in the Materials and methods section.
3.2. Effects of celecoxib on the sulfonation of 17β-E2 and its analogues with human liver cytosol
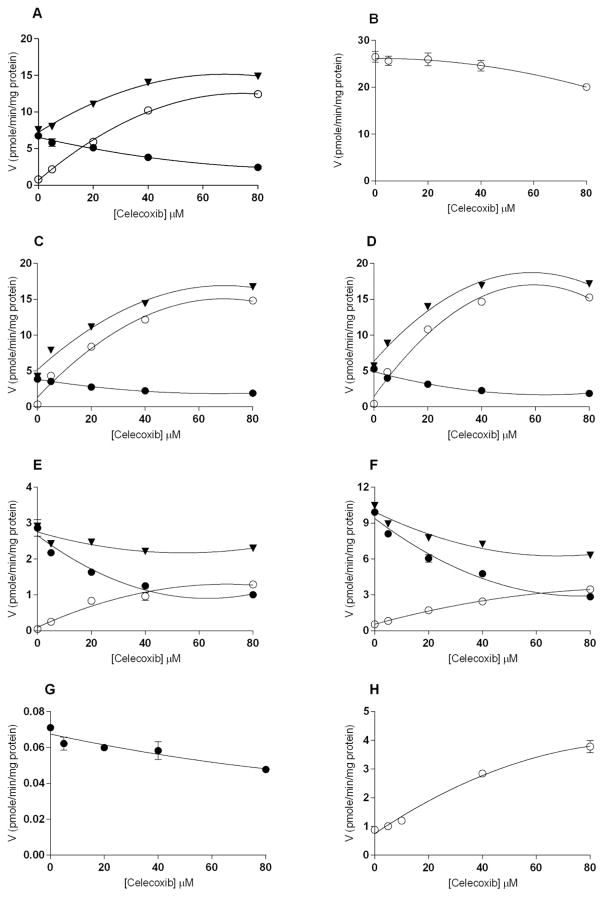
In the presence of celecoxib, the total production of 17β-E2 sulfates was stimulated at low concentrations of celecoxib (5–40 μM) and reached a plateau between 40 and 80 μM celecoxib (Figure 4A). The product ratio of 3-sulfate/17-sulfate reached 1 at approximately 20 μM celecoxib with human liver cytosol. At 40 μM celecoxib, there was 13-fold stimulation of 17β-E2–17-sulfate formation compared to incubations without celecoxib. When human liver cytosol was incubated with 0.4 μM 17α-E2 in the absence of celecoxib, the 17-sulfate was the predominant product (activity was 25.8 pmol/min/mg protein), with undetectable 3-sulfate activity. In the presence of celecoxib, a 20% inhibition was observed for the formation of the 17-sulfate at 80 μM (Figure 4B).
Figure 4.
Celecoxib affects the position of sulfonation of estradiol analogs in human liver cytosol. Graphs show the effects of celecoxib on the sulfonation of A) 17β-E2; B) 17α-E2; C) 6D-E2; D) 17β-Eqn; E) 9D-E2 F) 17β-Eq G) E1 and H) 3Me-E2 with human liver cytosol showing 3-sulfates (●), 17-sulfates (○) and total rate of sulfonation (▼). Sulfotransferase activity was assayed with varying concentrations of celecoxib in the presence of 0.4 μM of each substrate and 2 μM 35S-PAPS with human liver cytosol.
When using 6D-E2 (0.4 μM) as a substrate, in the presence of increased concentrations of celecoxib (2.5–80 μM), the 3-sulfate formation was reduced, while the 17-sulfate formation was greatly stimulated. As a result, the total product formation was stimulated (Figure 4C). The product ratio of 3-sulfate/17-sulfate reached 1 at 5 μM celecoxib with human liver cytosol. At 40 μM celecoxib, there was 35-fold stimulation of 17-sulfate formation relative to controls with no celecoxib.
The effects of celecoxib of 17β-Eqn sulfonation were similar to those seen with 6D-E2. The formation of 17-sulfate was greatly stimulated in the presence of celecoxib, but reached saturation at 40 μM celecoxib (Figure 4D). The product ratio of 3-sulfate/17-sulfate reached 1 at a celecoxib concentration of 5 μM. At 40 μM celecoxib, there was 34-fold stimulation of 17-sulfate formation.
When 9D-E2 was the substrate the generation of 17-sulfate was stimulated and that of 3-sulfate was inhibited, such that the ratio 9D-E2–17-sulfate/9D-E2–3-sulfate reached 1 at 40 μM celecoxib (Figure 4E). The stimulation of 17-sulfate was not large enough to increase the overall formation of sulfonated products.
The effect of celecoxib on 17β-Eq was similar to that on 9D-E2. When 17β-Eq was the substrate, although the amount of 17-sulfate product slowly increased in the presence of increasing concentrations of celecoxib, there was a greater decrease in 3-sulfate formation such that the total amount of sulfate product decreased with the increase in celecoxib concentration (Figure 4F). The product ratio of 3-sulfate/17-sulfate reached 1 at approximately 72 μM celecoxib with human liver cytosol.
With E1, which has no 17-hydroxy group, as substrate, 80 μM celecoxib inhibited sulfonation by 30% (Figure 4G). For the substrate with only a 17-β-hydroxy group, 3-Me-E2, adding up to 80 μM celecoxib to incubations with human liver cytosol resulted in increased sulfonation, with no apparent saturation of this effect over the studied celecoxib concentration range (Figure 4H). At 40 μM celecoxib, there was a 3.2-fold stimulation of 17-sulfate formation compared with controls without celecoxib.
3.3. Celecoxib inhibition and isozyme selectivity
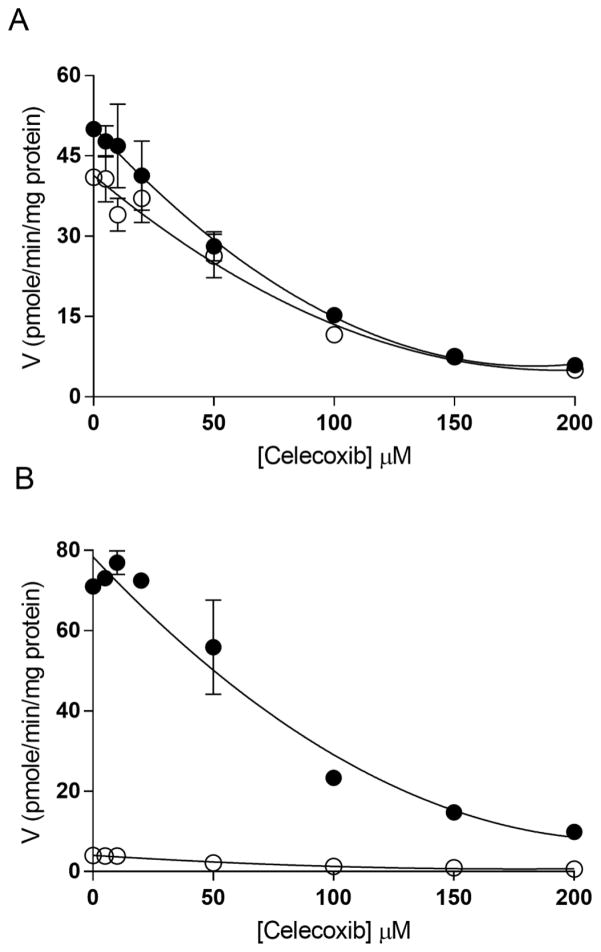
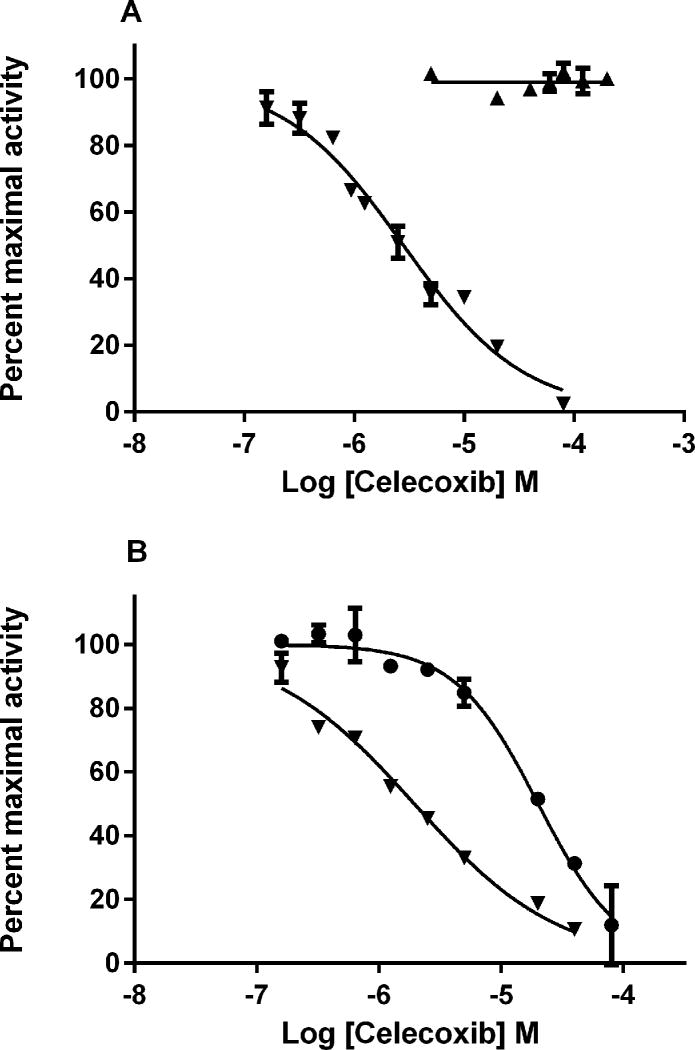
It was of interest to investigate the effects of celecoxib on other sulfotransferases, as the major phenol SULTs have been reported to sulfonate estrogens [8]. Among the isozymes tested SULT1B1 and 1A3 showed no activity towards 10 nM E1 (data not shown). Celecoxib did not inhibit E1 sulfonation by SULT1E1 significantly, but showed fairly potent inhibition of E1 sulfonation by SULT1A1 (IC50: 2.6 μM) (Figure 5A).
Figure 5.
Celecoxin inhibits the activity of SULT1A1 but not SULT1E1. Effects of celecoxib on the sulfonation of A) 10 nM E1 by SULT1A1 (▼), SULT1E1 (▲) and B) 4 μM pNP by SULT1A1 (▼) and human liver cytosol (●). Sulfotransferase activity was assayed with varying concentrations of celecoxib in the presence of 10 nM 3H-E1 and 20 μM PAPS with SULT1A1 (control activity: 2.7 pmol/min/mg protein) and SULT1E1 (control activity: 38.4 pmol/min/mg protein), and of 4 μM 14C-pNP and 20 μM PAPS with human liver cytosol (control activity: 5.4 nmol/min/mg protein) and SULT1A1 (control activity: 8.2 nmol/min/mg protein).
Sulfonation of 4 μM pNP was inhibited in human liver cytosol with IC50 of 20 μM. Human recombinant SULT1A1 was also sensitive to inhibition by celecoxib with an IC50 of 2.1 μM (Figure 5B).
3.4. Celecoxib-effect on 17β-E2 sulfonation by human liver cytosol in the presence of isozyme selective inhibitors
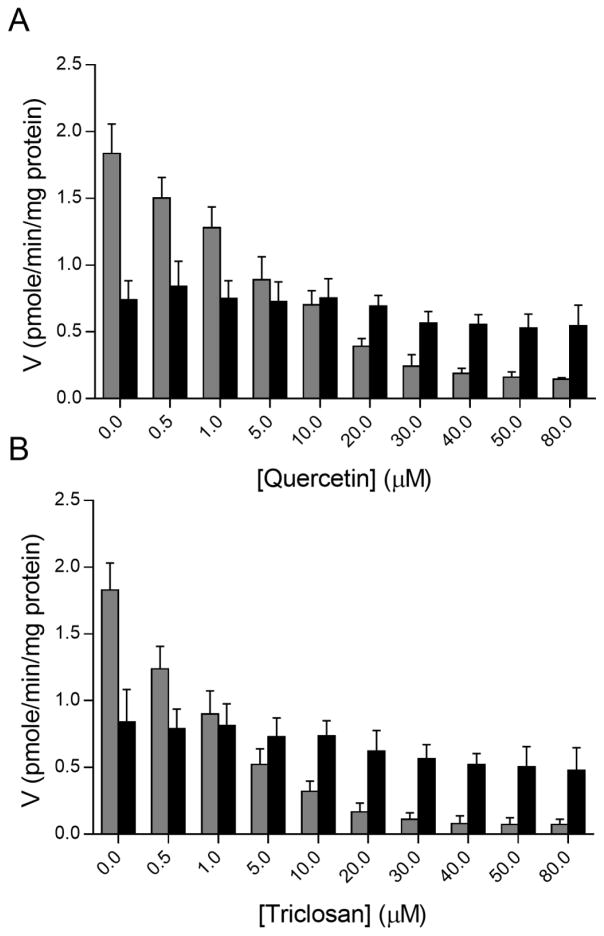
Selective inhibitors of phenol SULTs, quercetin and triclosan were incubated with human liver cytosol, 30 μM celecoxib and 50 nM 17β-E2. The concentration of celecoxib selected was previously shown to generate readily quantifiable amounts of both 17β-E2-17 sulfate and 3-sulfate. Quercetin and triclosan both inhibited 17β-E2–3-sulfate formation with IC50 values of 4 and 1 μM, respectively (Figure 6A and 6B). Quercetin did not significantly affect the formation of 17β-E2–17-sulfate in the range of concentrations studied, however triclosan concentrations greater than 30 μM caused a slight decrease in 17β-E2–17-sulfate formation.
Figure 6.
Inhibition of 17β-E2–3-sulfate formation by selective inhibitors of SULT1A1 and SULT1E1 in pooled human liver cytosol in the presence of celecoxib. (A) Quercetin (B) Triclosan. 17β-E2–3-sulfate is shown in grey and 17β-E2–17-sulfate is shown in black columns. The substrate concentration was 50 nM and the reaction mixtures were prepared as described in the Materials and methods section.
3.5. Celecoxib modulation – Human Rat Comparison
Liver cytosol from the rat, a common model animal, was tested for its capacity to sulfonate selected steroids and for the effect of celecoxib. As expected, sexual dimorphism was evident in the pattern of sulfonation of the steroids tested.
DHEA and AD were the most active substrates for female rat liver cytosol with control activities of 75.4 and 90.5 pmoles/min/mg protein, respectively (Table 1). They were followed by Epi-T and T which were less active than DHEA and AD (control activities are 21.5 and 25.0 pmoles/min/mg protein, respectively). In male rats, only DHEA and AD were sulfonated to any significant extent with control activities of 26.1 and 18.9 pmoles/min/mg protein, respectively. In the presence of 100 μM celecoxib the activities derived from both female and male rat were inhibited towards these non-aromatic steroids by 44–68% (Table 1). With human liver cytosol, the order of activities for the non-aromatic steroids was Epi-T≈DHEA≈AD > T, and addition of 80 μM celecoxib to incubations inhibited activity by 72–67% (Table 1).
Table 1.
Comparison of celecoxib’s effect on steroid sulfonation in pooled human liver cytosol, female and male rat liver cytosols. Each steroid was present at 400 nM. Activities are pmoles/min/mg protein and given as mean values for duplicate measurements in human and as mean ± SD for duplicate measurements in three each female and male rats.
|
Human liver cytosola
|
Female rat liver cytosolb
|
Male rat liver cytosolb
|
||||
|---|---|---|---|---|---|---|
| Control activity | Activity with Celecoxibc | Control activity | Activity with Celecoxibc | Control activity | Activity with Celecoxibc | |
| DHEA–S | 50.2 | 15.3 | 75.4 ±18.9 | 28.9 ± 6.21 | 26.1 ± 3.4 | 13.5 ± 5.2 |
| AD–S | 40.9 | 11.6 | 90.5 ± 22.1 | 30.2 ± 10.7 | 18.9 ± 2.1 | 10.5 ± 1.5 |
| Epi-T–S | 71.0 | 23.4 | 21.5 ± 4.0 | 8.1 ± 4.1 | ns | ns |
| T–S | 3.9 | 1.3 | 25.0 ± 4.6 | 9.4 ± 3.8 | ns | ns |
| 17β-E2–3S | 6.8 | 2.4 | 6.2 ± 0.8 | 4.5 ± 1.2 | 20.4 ± 4.3 | 6.1 ± 1.5 |
| 17β-E2–17S | 0.8 | 12.4 | 28.5 ± 4.5 | 24.1 ± 1.9 | 1.5 ± 0.5 | 0.4 ± 0.3 |
| 17α-E2–3S | ns | ns | ns | ns | 34.8 ± 8.5 | 8.9 ± 4.1 |
| 17α-E2–17S | 25.8 | 20.0 | 20.7 ± 6.8 | 6.8 ± 2.4 | ns | ns |
ns: No quantifiable sulfonation observed
The enzyme source is pooled human liver cytosol from three individual livers
Liver cytosol from three different rats was used as enzyme source
Activities in the presence of 100 μM celecoxib (80 μM celecoxib for 17β-E2 and 17α-E2 sulfates with human liver cytosol).
With 50 nM 17β-E2 as substrate, female rat liver cytosol generated predominantly the 17-sulfate (activity = 28.5 pmoles/min/mg protein), and the 3-sulfate (activity = 6.2 pmole/min/mg protein) was the minor product. This is in contrast to the human and male rat liver cytosol fractions, which had 3-sulfate as their major products (Table 1). In the presence of 80 and 100 μM celecoxib, the 3-sulfate generation by human and male rat liver cytosols was inhibited by 63% and 70%, respectively. Celecoxib was much less inhibitory for female rat liver cytosol (14% inhibition of 17-sulfate generation and 28% inhibition of 3-sulfate formation with 100 μM celecoxib). The most pronounced difference between the species was that the stimulation of 17-sulfate production by celecoxib observed with human liver was not seen for either of the rat sexes, instead celecoxib inhibited 17-sulfate formation. Another difference was that with 0.4 μM 17α-E2 as the substrate, human liver cytosol and female rat liver cytosol formed only 17-sulfate. However, with male rat liver cytosol 17α-E2 predominantly formed the 3-sulfate along with a barely detectable amount of 17-sulfate. Celecoxib (80–100 μM) inhibited the sulfonation of 17α-E2 in both human and rat liver cytosol (Table 1).
3.6. In silico studies
Celecoxib could dock in the PAPS binding site in addition to the substrate binding site of all SULTs tested i.e. human SULT1A1, 1E1, 2A1 and rat ST-60. The mechanistic implications of this binding to the PAPS site could be inhibition of enzyme activity, depending on the ease and conformation of substrate binding.
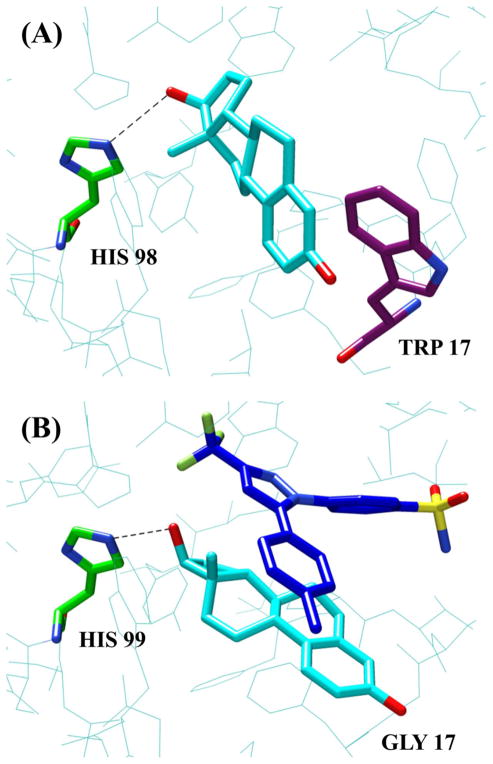
Ligand docking studies conducted on the homology model of ST-60, the most abundant form of hydroxysteroid sulfotransferase in female rat liver, showed that the favorable binding mode of 17β-E2 had the 17-hydroxy group of the ligand in hydrogen bonding distance to His-98, the catalytic residue required for sulfotransferase activity (Figure 7A). The model did not accommodate any satisfactory bindings of celecoxib as the volume of the ST-60 binding pocket is reduced compared to SULT2A1 because the side chain of Trp-17, which replaces Gly-17 in ST-60, reduced the volume of the binding pocket.
Figure 7.
Comparison of human SULT2A1 and its female rat analog, ST-60. (A) Binding site of ST-60 homology model docked with 17β-E2. (B) SULT2A1 binding site with docked celecoxib (blue stick model) and 17β-E2. The 17β-E2 models in both parts of the figure are shown as cyan sticks and the catalytic histidine residues shown as green sticks. Trp16 of ST-60, the amino acid possibly responsible for 17-sulfonation of 17β-E2 in ST-60 is shown as purple stick model.
4. DISCUSSION
Interaction of celecoxib with human liver cytosol resulted in modulation of the sulfonation of 17β-E2, DHEA and other physiologically important steroid and sterol substrates. This observation is consistent with our previous finding that celecoxib affected SULT2A1 enzyme activity towards these compounds [18], and the known expression pattern of SULT2A1 in human liver [7]. Celecoxib has been shown to stimulate 17-sulfate formation and inhibit 3-sulfate formation of 17β-E2 and its analogues, with SULT2A1 [18]. The structural requirements for this phenomenon appeared to be as follows: a 17β-hydroxy group and a free 3-phenolic hydroxy group, or an aromatic ring-A are essential; conjugated double bonds in ring-B decreased the formation of 3-sulfate, but stimulated the formation of 17-sulfate and increased the ratio of 17-sulfate to 3-sulfate. When human liver cytosol was used instead of SULT2A1, broadly similar effects of celecoxib on the sulfonation of 17β-E2 and its analogues were observed with some differences. These studies showed that for 17β-E2, 6D-E2, 17β-Eq 17β-Eqn and 9D-E2, addition of celecoxib to incubations resulted in switching of the dominant position of sulfonation. Examination of the ratio of 17-sulfate to 3-sulfate at increasing concentrations of celecoxib for these five steroids (measured at 0.4 μM substrate) revealed that 17β-Eqn was most sensitive to the effect on 17-sulfonation, followed in order by 6D-E2, 17β-E2, 17β-Eq and 9D-E2. As in the case of SULT2A1, stimulation of the formation of 17-sulfate of 3Me-E2 did not saturate in the range of celecoxib concentrations tested (0–80 μM) with pooled human liver cytosol. For 17β-Eq, celecoxib did not change 3-sulfate formation between 1.25 and 20 μM celecoxib with SULT2A1, but with liver cytosol, the rate of 3-sulfate formation was slightly decreased. This may be due to inhibition of SULT1A1-catalyzed 3-sulfonation of 17β-Eq by celecoxib in human liver cytosol. Our previous study showed that the 17-sulfate of 17α-E2 was the only sulfated product with SULT2A1 [18]. Similarly, when human liver cytosol was used in this study, 17-sulfate was the predominant product with 38-fold less 3-sulfate, and the addition of celecoxib did not affect the product profile. It is known that the 3-sulfate of 17β-E2 is the major product formed by human liver cytosol and that the phenol SULTs 1E1 and 1A1 are the major active enzymes forming this metabolite at low 17β-E2 concentrations [8]. The low rates of formation of 17α-E2–3-sulfate with human liver cytosol suggest that 17α-E2 is not a good substrate for SULT1E1 or SULT1A1. Thus, the change of configuration of the 17-hydroxy group alters the binding of 17α-E2 to SULT1E1, 1A1 or 2A1 in such a way that celecoxib has little effect on activity.
Celecoxib was a weak inhibitor of E1 sulfonation with human liver cytosol, with an IC50 value greater than 80 μM (Figure 4). The three enzymes expected to contribute to E1 sulfonation in liver, SULT1E1, SULT1A1 and SULT2A1, were affected differently by celecoxib. As reported in a previous paper, celecoxib did not affect the sulfonation of E1 with SULT2A1 [18]. As shown in Figure 5A, celecoxib inhibited E1-sulfonation with SULT1A1 with an IC50 value of 2.6 μM and had no effect on SULT1E1. These results suggest that celecoxib acts as a selective inhibitor of SULT1A1. It has been reported that pNP at 4 μM is a diagnostic substrate of SULT1A1 [30, 35]. Our results showed that celecoxib inhibited pNP sulfonation with recombinant SULT1A1 at a potency similar to that found with E1. Since celecoxib only weakly inhibited E1 sulfonation in human liver cytosol (Figure 4G), these results suggest that SULT1A1 plays a relatively minor role in E1 sulfonation, at least at the 10 nM substrate concentration studied.
When using expressed SULT2A1, it was shown that the 17β-E2–17-sulfate/17β-E2–3-sulfate ratio could reach 15 at 160 μM celecoxib, while the highest ratio was around 1 when using human liver cytosol [8]. It was proposed [8] that this is because SULT1E1 and SULT1A1, which form only the 3-sulfate, contribute to the sulfonation of 50 nM 17β-E2 in human liver cytosol. The phenol-type SULT1E1 and SULT1A1 rather than SULT2A1 are readily inhibited by xenobiotics [20, 21]. Quercetin, one of the most abundant flavonoids in vegetables, fruit and wine, is a more potent inhibitor of SULT1A1 than SULT1E1 with IC50 values ranging from pM to nM when using different substrates [20, 21]. It was reported that quercetin inhibited the sulfonation of 17β-E2 by rat SULT1A1 (17β-E2 concentration was 25 μM) and human SULT1E1 (17β-E2 concentration was 20 nM), with IC50 values of 0.29 μM and 1.4 μM, respectively [36]. Triclosan showed more potent inhibition of SULT1E1 with 1 nM 17β-E2 as the substrate (IC50 = 27 nM, unpublished) than that of SULT1A1 with 4 μM pNP as the substrate (IC50 = 3.6 μM) [24]. The potent inhibition of 3-sulfate formation by triclosan in the presence of celecoxib (with IC50 value of 1 μM) and less potent inhibition by quercetin (IC50 value of 4 μM) further suggests that the isozymes that contribute to formation of 17β-E2-3-sulfate were mainly SULT1E1 and SULT2A1, with some contribution from SULT1A1.
Many xenobiotics, including environmental and dietary chemicals, are potent inhibitors of phenol SULTs [20, 21]. Preferential inhibition of 3-sulfate formation and stimulation of 17-sulfate formation suggested that the 17-sulfate/3-sulfate ratio would be increased in the presence of both phenol SULT inhibitors and celecoxib, and our results shown in Figure 6 confirm this. As the 17-sulfate is poorly converted to 17β-E2 by sulfatase, compared to 17β-E2–3-sulfate [16, 17], this interaction could be beneficial as it could lead to reduced 17β-E2 being available to target organs, such as the breast.
This work revealed considerable differences between rat and human in steroid sulfonation. The homology model of ST-60, which is the most abundant form of hydroxysteroid SULT in the female rat [26] is similar in amino acid composition and the overall structure of the binding site to the human SULT2A1 (Figure 7A). The amino acids His-99, Trp-77, Tyr-160, Trp-72, Pro-43, Phe-18, Pro-14 in SULT2A1 have identical counterparts in the structure of ST-60. The important differences are: Gly-17 of SULT2A1 is replaced by Trp-16, Met-16 by Phe-15 and the Tyr-238 by Leu-235. The difference of consequence for the species difference between female rat and human seems to be the replacement of Gly-17 in SULT2A1 by Trp-16 in ST-60, as the hydrophobic side chain of this tryptophan is inverted into the binding pocket, thereby truncating it. This truncation prohibits the binding of 17β-E2 with the 3-OH group within 3 Å of histidine 99, as found in SULT2A1, and allows only the orientation that has the 17-hydroxy group in the catalytic position. Trp-16 in ST-60 seems to be doing what celecoxib docking does to the binding site of SULT2A1, i.e. orienting the 17-OH group for sulfonation (Figure 7B).
5. CONCLUDING REMARKS
Over the past half-century, a growing belief among women and their physicians held that estrogen replacement therapy in postmenopausal women would prevent many of the manifestations of aging, including coronary heart disease (CHD), osteoporosis, and a decline in cognitive and sexual function. Premarin had been the most-prescribed drug in the United States to treat menopausal hot flashes, night sweats and vaginal dryness. Premarin contains 17β-E2, 17α-E2, 17β-Eq, 17β-Eqn along with other estrogens, mainly as in their sulfonated form [37]. However, multiple lines of evidence support an increased risk of breast cancer with estrogen use, including cell culture studies, [38], animal models, [39] and epidemiological studies [40–42]. In the Heart and Estrogen/progestin Replacement Study (HERS) and the Women’s Health Initiative studies, the risk of breast cancer in postmenopausal women was increased about 25% in those taking estrogen plus progestin [43–45]. Co-administration of celecoxib may reduce the risk of breast cancer incidence in postmenopausal women based on the following speculations: 1) E1–sulfate is the major component in Premarin, and it is metabolized to E1 and 17β-E2 [46]. It is well accepted that 17β-E2 is one of the most important factors in the growth and evolution of hormone-dependent breast tumors. E1, after conversion to 17β-E2, increases the amount of progesterone receptors and pS2 protein, as well as E1-inducible protein cathepsin D in hormone-dependent breast cancer cells (MCF-7) [47–51]. In breast tumors, in vivo and in vitro studies show that the preferential conversion is the reduction of E1 to 17β-E2 [52, 53]. With the presence of celecoxib, the major 17β-E2 sulfate product was switched from 3-sulfate to 17-sulfate, which is not readily hydrolyzed by sulfatase in breast cells, and therefore, the 17β-E2 levels will be reduced. 2) Celecoxib switches the sulfate products of 17β-Eq and 17β-Eqn from 3-sulfates to 17-sulfates. Similar to 17β-E2–17-sulfate, these 17-sulfates do not cause any biological response on estrogen and progesterone receptors [47, 48]. 3) Equilin and equilenin are transformed to 17β-Eq and 17β-Eqn in breast cancer cells [54] and normal postmenopausal women as well as in men [55]. Celecoxib may help reduce the 17β-E2 levels and inactivate the equine estrogens in breast cells by switching the 3-sulfates to 17-sulfates, thus possibly lowering the incidence of breast cancer.
In summary, this paper presents our findings that celecoxib interacts with several SULTs. Celecoxib changed the dominant position of sulfonation of 17β-E2 and its analogues including 6D-E2, 17β-Eqn,17β-Eq and 9D-E2, but not 17α-E2 with human liver cytosol, due to the presence of SULT2A1. Celecoxib was also able to stimulate the overall sulfonation of 3Me-E2, 17β-E2, 6D-E2, 17β-Eqn, 17β-Eq and 9D-E2. SULT inhibitors such as quercetin and triclosan suppressed the formation of 17β-E2–3-sulfate, while the formation of 17-sulfate was stimulated in the presence of celecoxib at low concentrations of SULT inhibitors. Celecoxib acts as a selective inhibitor of SULT1A1 with E1 and pNP as substrates. Neither male nor female rats are good models for studying the effect of celecoxib on human 17β-E2 sulfonation, however the structural comparison between human SULT2A1 and its analog in female rat liver shows that celecoxib in SULT2A1 may be assuming the role of Trp-16 in the rat enzyme in directing sulfonation of 17β-E2 to the 17-position.
HIGHLIGHTS.
Celecoxib inhibits SULT1A1 activity but not that of SULT1E1
Celecoxib affects sulfonation of physiologically important steroids in human liver
17-sulfonation of equine estrogens is stimulated by celecoxib in human liver
Effects of celecoxib on steroid sulfonation in rat liver differ from human liver
Acknowledgments
This study was supported in part by the grant R03CA123575 from the US Public Health Service and by the National Institutes of Health (NIH) and National Center for Research Resources (NCRR) CTSA grant UL1 TR000064 (Clinical and Translational Science Award). The authors would like to thank Dr. Charles N. Falany (University of Alabama at Birmingham, AL) for providing the hSULT2A1 expression system.
ABBREVIATIONS
- 17β-E2
17β-estradiol
- 17S
17-sulfate
- 17α-E2
17-α-estradiol
- 3Me-E2
3-methyl ether of estradiol
- 3S
3-sulfate
- 6D-E2
6-dehydroestradiol
- 9D-E2
9-dehydroestradiol
- AD
5-androsten-3β,17β-diol
- COX-2
cyclooxygenase-2
- DHEA
dehydroepiandrosterone
- DMSO
dimethyl sulfoxide
- Epi-T
epitestosterone
- 17β-Eq
17β-dihydroequilin
- 17β-Eqn
17β-dihydroequilenin
- IC50
concentration required to achieve 50% inhibition
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- PDB
protein data bank
- SULT
sulfotransferase
- T
testosterone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Coughtrie MW. Sulphation catalysed by the human cytosolic sulphotransferases--chemical defence or molecular terrorism? Hum Exp Toxicol. 1996;15(7):547–555. doi: 10.1177/096032719601500701. [DOI] [PubMed] [Google Scholar]
- 2.Strott CA. Sulfonation and molecular action. Endocr Rev. 2002;23(5):703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 3.Otake Y, Nolan AL, Walle UK, Walle T. Quercetin and resveratrol potently reduce estrogen sulfotransferase activity in normal human mammary epithelial cells. J Steroid Biochem Mol Biol. 2000;73(5):265–270. doi: 10.1016/s0960-0760(00)00073-x. [DOI] [PubMed] [Google Scholar]
- 4.Pasqualini JR, Chetrite GS. Estrone sulfatase versus estrone sulfotransferase in human breast cancer: potential clinical applications. J Steroid Biochem Mol Biol. 1999;69(1–6):287–292. doi: 10.1016/s0960-0760(99)00082-5. [DOI] [PubMed] [Google Scholar]
- 5.Pasqualini JR, Chetrite GS. Recent insight on the control of enzymes involved in estrogen formation and transformation in human breast cancer. J Steroid Biochem Mol Biol. 2005;93(2–5):221–236. doi: 10.1016/j.jsbmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5(5):e97. doi: 10.1371/journal.pbio.0050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riches Z, Stanley EL, Bloomer JC, Coughtrie MW. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab Dispos. 2009;37(11):2255–2261. doi: 10.1124/dmd.109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang LQ, James MO. Sulfotransferase 2A1 forms estradiol-17-sulfate and celecoxib switches the dominant product from estradiol-3-sulfate to estradiol-17-sulfate. J Steroid Biochem Mol Biol. 2005;96(5):367–374. doi: 10.1016/j.jsbmb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Kim B, Giardiello FM. Chemoprevention in familial adenomatous polyposis. Best Pract Res Clin Gastroenterol. 2011;25(4–5):607–622. doi: 10.1016/j.bpg.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu GD, Pathangey LB, Tinder TL, Lagioia M, Gendler SJ, Mukherjee P. Cyclooxygenase-2 inhibitor induces apoptosis in breast cancer cells in an in vivo model of spontaneous metastatic breast cancer. Mol Cancer Res. 2004;2(11):632–642. [PubMed] [Google Scholar]
- 11.Levitt RJ, Buckley J, Blouin MJ, Schaub B, Triche TJ, Pollak M. Growth inhibition of breast epithelial cells by celecoxib is associated with upregulation of insulin-like growth factor binding protein-3 expression. Biochem Biophys Res Commun. 2004;316(2):421–428. doi: 10.1016/j.bbrc.2004.02.062. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Lawson KA, Simmons-Menchaca M, Sun L, Sanders BG, Kline K. Vitamin E analog alpha-TEA and celecoxib alone and together reduce human MDA-MB-435-FL-GFP breast cancer burden and metastasis in nude mice. Breast Cancer Res Treat. 2004;87(2):111–121. doi: 10.1023/B:BREA.0000041593.69178.57. [DOI] [PubMed] [Google Scholar]
- 13.Guastalla JP, Bachelot T, Ray-Coquard I. Cyclooxygenase 2 and breast cancer. From biological concepts to therapeutic trials. Bull Cancer. 2004;91(Spec No):S99–108. [PubMed] [Google Scholar]
- 14.Brandao RD, Veeck J, Van de Vijver KK, Lindsey P, de Vries B, van Elssen CH, Blok MJ, Keymeulen K, Ayoubi T, Smeets HJ, Tjan-Heijnen VC, Hupperets PS. A randomised controlled phase II trial of pre-operative celecoxib treatment reveals anti-tumour transcriptional response in primary breast cancer. Breast Cancer Res. 2013;15(2):R29. doi: 10.1186/bcr3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arun B, Goss P. The role of COX-2 inhibition in breast cancer treatment and prevention. Semin Oncol. 2004;31(2 Suppl 7):22–29. doi: 10.1053/j.seminoncol.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 16.Chetrite GS, Cortes-Prieto J, Philippe JC, Wright F, Pasqualini JR. Comparison of estrogen concentrations, estrone sulfatase and aromatase activities in normal, and in cancerous, human breast tissues. J Steroid Biochem Mol Biol. 2000;72(1–2):23–27. doi: 10.1016/s0960-0760(00)00040-6. [DOI] [PubMed] [Google Scholar]
- 17.Pasqualini JR, Gelly C, Nguyen BL, Vella C. Importance of estrogen sulfates in breast cancer. J Steroid Biochem. 1989;34(1–6):155–163. doi: 10.1016/0022-4731(89)90077-0. [DOI] [PubMed] [Google Scholar]
- 18.Ambadapadi S, Wang PL, Palii SP, James MO. Celecoxib influences steroid sulfonation catalyzed by human recombinant sulfotransferase 2A1. J Steroid Biochem Mol Biol. 2015;152:101–113. doi: 10.1016/j.jsbmb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhavnani BR, Stanczyk FZ. Pharmacology of conjugated equine estrogens: efficacy, safety and mechanism of action. J Steroid Biochem Mol Biol. 2014;142:16–29. doi: 10.1016/j.jsbmb.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Wang LQ, James MO. Inhibition of sulfotransferases by xenobiotics. Curr Drug Metab. 2006;7(1):83–104. doi: 10.2174/138920006774832596. [DOI] [PubMed] [Google Scholar]
- 21.James MO, Ambadapadi S. Interactions of cytosolic sulfotransferases with xenobiotics. Drug Metab Rev. 2013;45(4):401–414. doi: 10.3109/03602532.2013.835613. [DOI] [PubMed] [Google Scholar]
- 22.De Santi C, Pietrabissa A, Mosca F, Rane A, Pacifici GM. Inhibition of phenol sulfotransferase (SULT1A1) by quercetin in human adult and foetal livers. Xenobiotica. 2002;32(5):363–368. doi: 10.1080/00498250110119108. [DOI] [PubMed] [Google Scholar]
- 23.Marchetti F, De Santi C, Vietri M, Pietrabissa A, Spisni R, Mosca F, Pacifici GM. Differential inhibition of human liver and duodenum sulphotransferase activities by quercetin, a flavonoid present in vegetables, fruit and wine. Xenobiotica. 2001;31(12):841–847. doi: 10.1080/00498250110069159. [DOI] [PubMed] [Google Scholar]
- 24.Wang LQ, Falany CN, James MO. Triclosan as a substrate and inhibitor of 3′-phosphoadenosine 5′-phosphosulfate-sulfotransferase and UDP-glucuronosyl transferase in human liver fractions. Drug Metab Dispos. 2004;32(10):1162–1169. doi: 10.1124/dmd.104.000273. [DOI] [PubMed] [Google Scholar]
- 25.James MO, Li W, Summerlot DP, Rowland-Faux L, Wood CE. Triclosan is a potent inhibitor of estradiol and estrone sulfonation in sheep placenta. Environ Int. 2010;36(8):942–949. doi: 10.1016/j.envint.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Klaassen CD. Ontogeny and hormonal basis of female-dominant rat hepatic sulfotransferases. J Pharmacol Exp Ther. 1996;279(1):386–391. [PubMed] [Google Scholar]
- 27.Liu L, Klaassen CD. Ontogeny and hormonal basis of male-dominant rat hepatic sulfotransferases. Mol Pharmacol. 1996;50(3):565–572. [PubMed] [Google Scholar]
- 28.Prather B, Ethen CM, Machacek M, Wu ZL. Golgi-resident PAP-specific 3′-phosphatase-coupled sulfotransferase assays. Anal Biochem. 2012;423(1):86–92. doi: 10.1016/j.ab.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Wang LQ, Lehmler HJ, Robertson LW, Falany CN, James MO. In vitro inhibition of human hepatic and cDNA-expressed sulfotransferase activity with 3-hydroxybenzo[a]pyrene by polychlorobiphenylols. Environ Health Perspect. 2005;113(6):680–687. doi: 10.1289/ehp.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coughtrie MW, Johnston LE. Interactions between dietary chemicals and human sulfotransferases-molecular mechanisms and clinical significance. Drug Metab Dispos. 2001;29(4 Pt 2):522–528. [PubMed] [Google Scholar]
- 31.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 32.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37(Database issue):D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38(3):305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 35.Cappiello M, Giuliani L, Pacifici GM. Differential distribution of phenol and catechol sulphotransferases in human liver and intestinal mucosa. Pharmacology. 1990;40(2):69–76. doi: 10.1159/000138643. [DOI] [PubMed] [Google Scholar]
- 36.Mesia-Vela S, Kauffman FC. Inhibition of rat liver sulfotransferases SULT1A1 and SULT2A1 and glucuronosyltransferase by dietary flavonoids. Xenobiotica. 2003;33(12):1211–1220. doi: 10.1080/00498250310001615762. [DOI] [PubMed] [Google Scholar]
- 37.Bhavnani BR. Estrogens and menopause: pharmacology of conjugated equine estrogens and their potential role in the prevention of neurodegenerative diseases such as Alzheimer’s. J Steroid Biochem Mol Biol. 2003;85(2–5):473–482. doi: 10.1016/s0960-0760(03)00220-6. [DOI] [PubMed] [Google Scholar]
- 38.Fuchs-Young R, Glasebrook AL, Short LL, Draper MW, Rippy MK, Cole HW, Magee DE, Termine JD, Bryant HU. Raloxifene is a tissue-selective agonist/antagonist that functions through the estrogen receptor. Ann N Y Acad Sci. 1995;761:355–360. doi: 10.1111/j.1749-6632.1995.tb31392.x. [DOI] [PubMed] [Google Scholar]
- 39.Clemons M, Goss P. Estrogen and the risk of breast cancer. N Engl J Med. 2001;344(4):276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 40.Beral V. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2003;362(9382):419–427. doi: 10.1016/s0140-6736(03)14065-2. [DOI] [PubMed] [Google Scholar]
- 41.C.G.o.H.F.i.B. Cancer. Breast cancer and hormone replacement therapy: collaborative reanalysis of data from 51 epidemiological studies of 52,705 women with breast cancer and 108,411 women without breast cancer. Collaborative Group on Hormonal Factors in Breast Cancer. Lancet. 1997;350(9084):1047–1059. [PubMed] [Google Scholar]
- 42.Crosignani PG. Breast cancer and hormone-replacement therapy in the Million Women Study. Maturitas. 2003;46(2):91–92. doi: 10.1016/j.maturitas.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Hulley S, Grady D, Bush T, Furberg C, Herrington D, Riggs B, Vittinghoff E. Randomized trial of estrogen plus progestin for secondary prevention of coronary heart disease in postmenopausal women. Heart and Estrogen/progestin Replacement Study (HERS) Research Group. Jama. 1998;280(7):605–613. doi: 10.1001/jama.280.7.605. [DOI] [PubMed] [Google Scholar]
- 44.Rossouw JE, Anderson GL, Prentice RL, LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SA, Howard BV, Johnson KC, Kotchen JM, Ockene J. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results From the Women’s Health Initiative randomized controlled trial. Jama. 2002;288(3):321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 45.Simon JA, Hsia J, Cauley JA, Richards C, Harris F, Fong J, Barrett-Connor E, Hulley SB. Postmenopausal hormone therapy and risk of stroke: The Heart and Estrogen-progestin Replacement Study (HERS) Circulation. 2001;103(5):638–642. doi: 10.1161/01.cir.103.5.638. [DOI] [PubMed] [Google Scholar]
- 46.Ruder HJ, Loriaux L, Lipsett MB. Estrone sulfate: production rate and metabolism in man. J Clin Invest. 1972;51(4):1020–1233. doi: 10.1172/JCI106862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasqualini JR, Gelly C, Lecerf F. Biological effects and morphological responses to estriol, estriol-3-sulfate, estriol-17-sulfate and tamoxifen in a tamoxifen-resistant cell line (R-27) derived from MCF-7 human breast cancer cells. Eur J Cancer Clin Oncol. 1986;22(12):1495–1501. doi: 10.1016/0277-5379(86)90086-6. [DOI] [PubMed] [Google Scholar]
- 48.Pasqualini JR, Gelly C, Lecerf F. Estrogen sulfates: biological and ultrastructural responses and metabolism in MCF-7 human breast cancer cells. Breast Cancer Res Treat. 1986;8(3):233–240. doi: 10.1007/BF01807336. [DOI] [PubMed] [Google Scholar]
- 49.Santner SJ, Ohlsson-Wilhelm B, Santen RJ. Estrone sulfate promotes human breast cancer cell replication and nuclear uptake of estradiol in MCF-7 cell cultures. Int J Cancer. 1993;54(1):119–124. doi: 10.1002/ijc.2910540119. [DOI] [PubMed] [Google Scholar]
- 50.Vignon F, Terqui M, Westley B, Derocq D, Rochefort H. Effects of plasma estrogen sulfates in mammary cancer cells. Endocrinology. 1980;106(4):1079–1086. doi: 10.1210/endo-106-4-1079. [DOI] [PubMed] [Google Scholar]
- 51.Westley B, Rochefort H. A secreted glycoprotein induced by estrogen in human breast cancer cell lines. Cell. 1980;20(2):353–362. doi: 10.1016/0092-8674(80)90621-2. [DOI] [PubMed] [Google Scholar]
- 52.Miettinen M, Mustonen M, Poutanen M, Isomaa V, Wickman M, Soderqvist G, Vihko R, Vihko P. 17Beta-hydroxysteroid dehydrogenases in normal human mammary epithelial cells and breast tissue. Breast Cancer Res Treat. 1999;57(2):175–182. doi: 10.1023/a:1006217400137. [DOI] [PubMed] [Google Scholar]
- 53.Poutanen M, Moncharmont B, Vihko R. 17 beta-hydroxysteroid dehydrogenase gene expression in human breast cancer cells: regulation of expression by a progestin. Cancer Res. 1992;52(2):290–294. [PubMed] [Google Scholar]
- 54.Spink DC, Zhang F, Hussain MM, Katz BH, Liu X, Hilker DR, Bolton JL. Metabolism of equilenin in MCF-7 and MDA-MB-231 human breast cancer cells. Chem Res Toxicol. 2001;14(5):572–581. doi: 10.1021/tx000219r. [DOI] [PubMed] [Google Scholar]
- 55.Bhavnani BR, Cecutti A. Metabolic clearance rate of equilin sulfate and its conversion to plasma equilin, conjugated and unconjugated equilenin, 17 beta-dihydroequilin, and 17 beta-dihydroequilenin in normal postmenopausal women and men under steady state conditions. J Clin Endocrinol Metab. 1993;77(5):1269–1274. doi: 10.1210/jcem.77.5.8077320. [DOI] [PubMed] [Google Scholar]