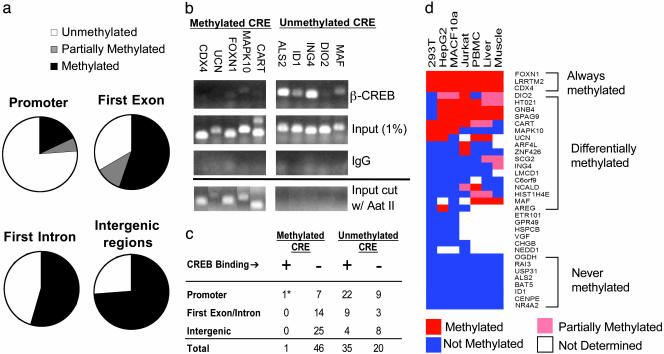
Fig. 2.
CRE methylation blocks CREB binding to nonfunctional sites. (a) CRE methylation frequency on promoters and exonic, intronic, or intergenic regions, as determined by endonuclease assay with methylation-sensitive restriction enzyme Aat II. (b) Effect of CRE methylation on CREB occupancy in HEK293T cells by ChIP assay. (Upper) Relative binding of CREB to unmethylated or methylated CREs in five different promoters for each group by using anti-CREB antiserum. Input levels of DNA (1%) for each gene are indicated; nonspecific IgG control is shown. (Lower) PCR analysis of genomic fragments showing that methylated CREs are resistant to Aat II digestion, whereas unmethylated CREs are completely digested. (c) Summary of CRE methylation and CREB-binding patterns in promoters, intergenic, or intronic/exonic regions of the genome. * marks one CREB-occupied CRE (LRRTM2 gene) that was partially methylated in HEK293T cells. (d) Relative methylation state (color-coded) of 34 genes across seven cell contexts. Genes are clustered based on methylation profiles.