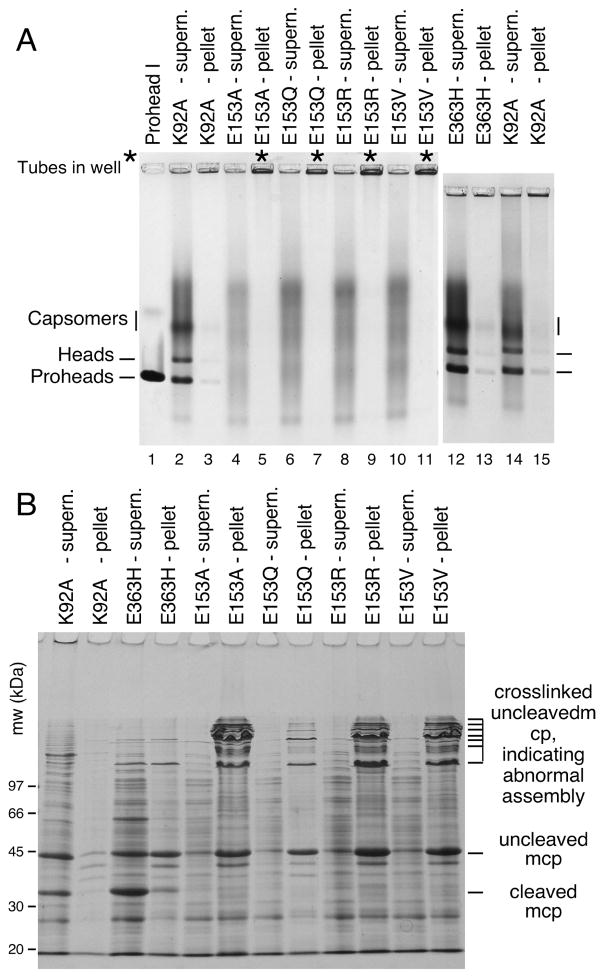
Figure 3. Mutants that disrupt the E153-R210 salt bridge assemble abnormally.
A. Agarose gel analysis. Crude supernatant (supern.), and pellet fractions from autoinduced lysates were run on a 0.9% agarose gel. The E153 mutants showed no distinct bands. Controls K92A and E363H each produced strong proheads and head bands, which are mixtures of Prohead I, Prohead II and the Head II produced by the freeze/thaw step during preparation. Asterisks (“*”) mark the stainable material that appeared in the wells. B. SDS gel analysis. Pellets and supernatants were denatured and analyzed on SDS gels. Control mutants K92A and E363H make Prohead I (PI) and Prohead II (PII), so they produce control bands of 42 kDa uncleaved major capsid protein (in PI) and of 31 kDa cleaved major capsid protein (in PII), and a ladder crosslinked bands induced by the freeze/thaw step during preparation. E153 mutants produced uncleaved major capsid protein and a ladder of high-molecular-weight bands (crosslinked oligomers of uncleaved major capsid protein that indicate that abnormal assembly has occurred).