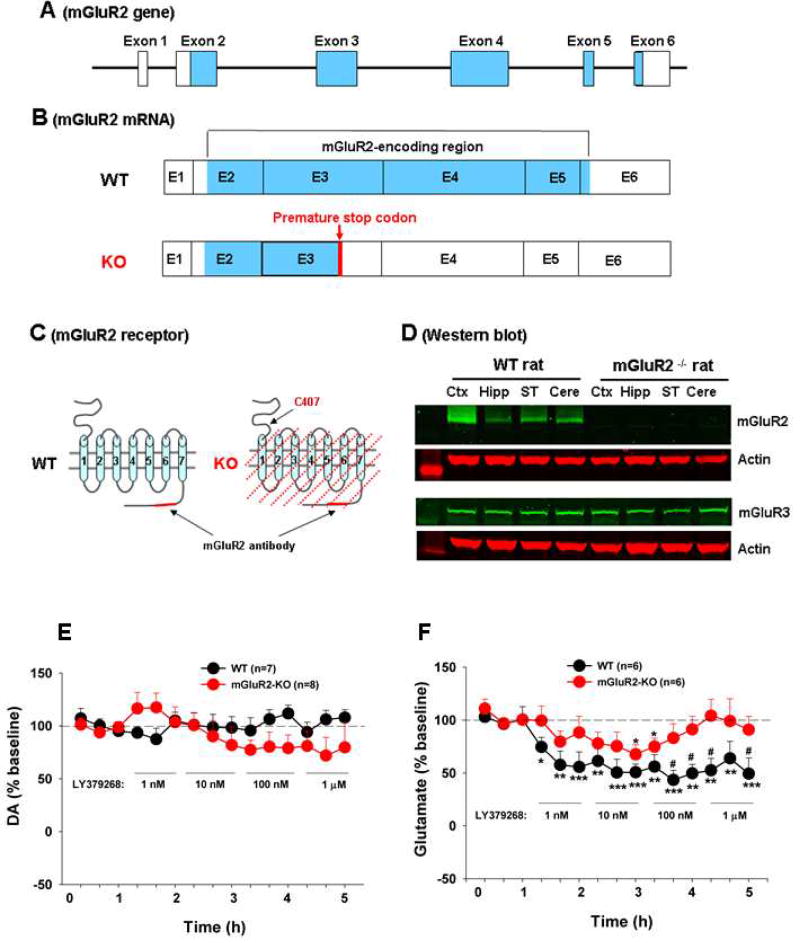
Figure 1.
mGluR2 receptor expression in WT and mGluR2-KO rats. (A) Rat mGluR2 genomic structure, illustrating the 6 exons and the mGluR2-encoding regions (blue color). (B) mGluR2 transcripts (mRNA) and the location of the premature stop codon that causes the termination of mGluR2 protein translation in mGluR2-knockout (mGluR2-KO) rats. (C) mGluR2 receptor structure in WT rats and the predicted receptor deletion region in mGluR2-KO rats. (D) Representative immunoblot bands, showing mGluR2 or mGluR3 expression in cerebral cortex (Ctx), hippocampus (Hipp), striatum (ST) and cerebellum (cere) of WT and mGluR2-KO rats. (E/F) In vivo microdialysis data, showing intra-NAc local perfusion of LY379268 failed to alter extracellular DA efflux (E), but significantly decreased extracellular glutamate levels in both WT and mGluR2-KO rats (F). However, the reduction in extracellular glutamate was significantly attenuated in mGluR2-KO rats than in WT rats. The data are represented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, compared to baseline before LY379268 administration. #p<0.05, compared to mGluR2-KO rats. Also see Figure S1.