Fig. 5. Segmental labeling of RNA.
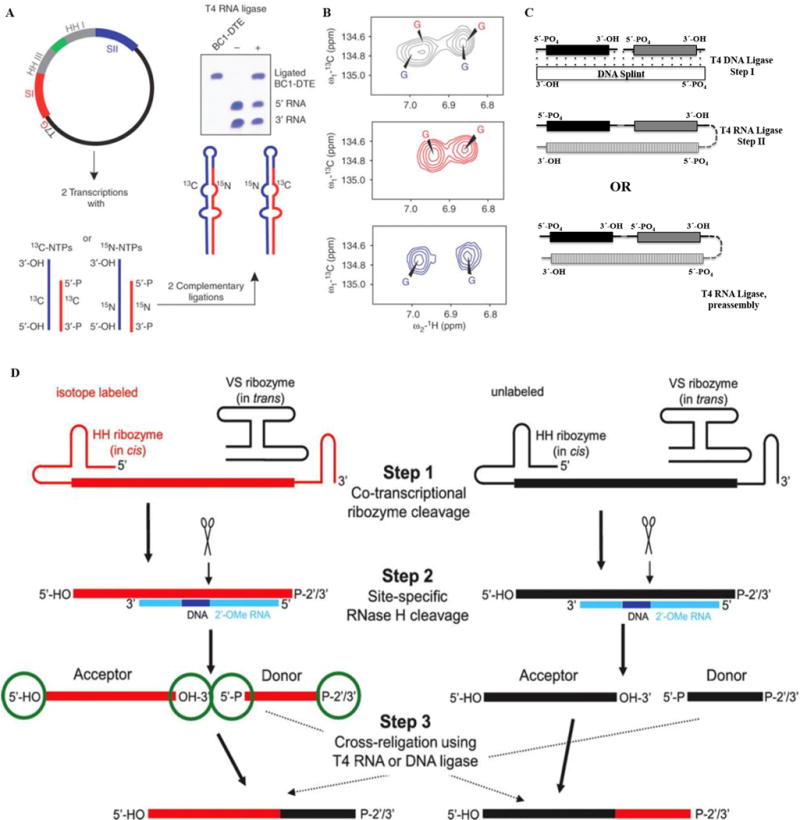
(A–B) Overview of the preparation of a segmentally isotope labeled RNA. Here (A) shows the construction of a plasmid where 3′-HH and 5′-HH are engineered. Panel (B) shows the simplification of aromatic regions of 1H, 13C-TROSY spectra for the RNA, the top panel shows spectra for uniformly 13C/15N-labeled RNA (black), middle for 5′13C- and 3′15N-labeled RNA (red) and the bottom spectrum corresponds to 5′15N- and 3′13C- labeled RNA (blue). The figure is taken from Tzakos et al. [54]. (C) Segmental labeling approach based on T4 DNA ligase, DNA splint and T4 RNA ligase adapted from Nelissen et al. [55]. The segmentally labeled RNA can be prepared either in single or multi-step ligations to obtain the desired product. Here, dark black, grey and vertically patterned fill colors represent three RNA fragments to be joined either in a two-step ligation based on T4 DNA ligase/DNA splint/T4 RNA ligase or single step ligation by T4 RNA ligase. Each fragment can be differently labeled or unlabeled. (D) A newly developed segmental labeling approach from Duss et al. [24] based on HH/VS ribozymes, 2′-O-methyl RNA/DNA chimera, RNase H and T4 DNA or T4 RNA ligases. In the first step, HH/VS ribozyme cleavage occurs co-transcriptionally, followed by site-specific RNase H cleavage in step II. T4 DNA/RNA ligase based ligation is done in step III. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
