Figure 2.
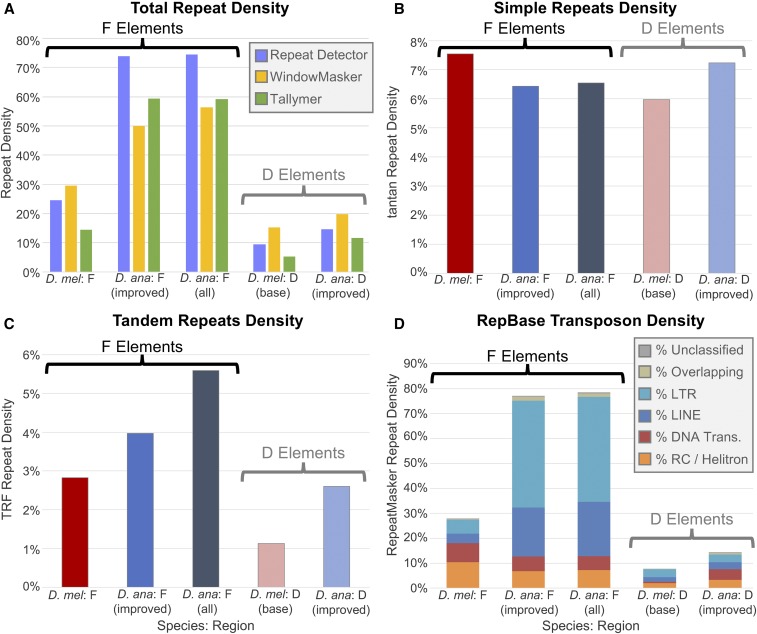
Expansion of the D. ananassae F element can primarily be attributed to the high density of LTR and LINE retrotransposons. (A) Total repeat density estimates from de novo repeat finders (Red, WindowMasker, and Tallymer) show that the D. ananassae F element has higher repeat density than the D. melanogaster F element (56.4–74.5% vs. 14.4–29.5%). Both F elements show higher repeat density than the euchromatic reference regions from the base of the D elements in D. ananassae (11.6–19.8%) and in D. melanogaster (5.4–15.2%). The repeat densities of the improved D. ananassae F-element scaffolds [D. ana: F (improved)] are similar to the repeat densities for all D. ananassae F-element scaffolds [D. ana: F (all)]. (B) Results from the tantan analysis show that the five analysis regions from D. melanogaster and D. ananassae have similar simple repeat density (6.0–7.5%). (C) TRF analysis shows that D. ananassae has higher tandem repeats density than D. melanogaster on both the F (5.6 vs. 2.8%) and the D elements (2.6 vs. 1.1%). (D) RepeatMasker analysis using the Drosophila RepBase library shows that the F element has higher transposon density than the D element both in D. melanogaster (28.0 vs. 7.7%) and in D. ananassae (78.6 vs. 14.4%). There is a substantial increase in the density of LTR and LINE retrotransposons on the D. ananassae F element compared to the D. melanogaster F element (42.1 vs. 5.5% and 21.8 vs. 3.8%, respectively). The D. ananassae euchromatic reference region also shows higher transposon density than D. melanogaster, but most of the difference can be attributed to the density of DNA transposons (4.3 vs. 0.6%). The region of overlap between two repeat fragments is classified as “Overlapping” if the two repeats belong to different repeat classes.