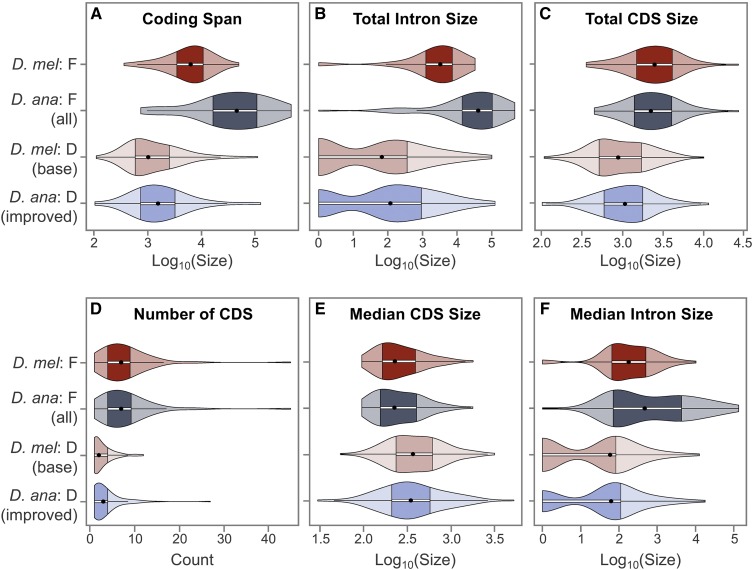
Figure 4.
F-element genes show distinct gene characteristics compared to D-element genes. Each violin plot is comprised of a box plot and a kernel density plot. The ● in each violin plot denotes the median and the darker region demarcates the IQR, which spans from the first (Q1) to the third (Q3) quartiles. The whiskers extending from the darker region spans from Q1 = −1.5 × IQR to Q3 = +1.5 × IQR; data points beyond the whiskers are classified as outliers. (A) D. ananassae F-element genes have larger coding spans (start codon to stop codon, including introns) than D. melanogaster F-element genes. (B) The D. ananassae F-element genes have larger coding spans because they have larger total intron sizes than D. melanogaster F-element genes. (C) F-element genes have larger total coding exon (CDS) sizes than D-element genes in both D. ananassae and D. melanogaster. (D) F-element genes have more CDS than D-element genes. (E) F-element genes have smaller median CDS size than D-element genes. (F) The median intron size for D. ananassae and D. melanogaster F-element genes shows a bimodal distribution; this distribution pattern indicates that the expansion of the coding spans of D. ananassae F-element genes compared to D. melanogaster F-element genes can be attributed to the substantial expansion of a subset of introns.