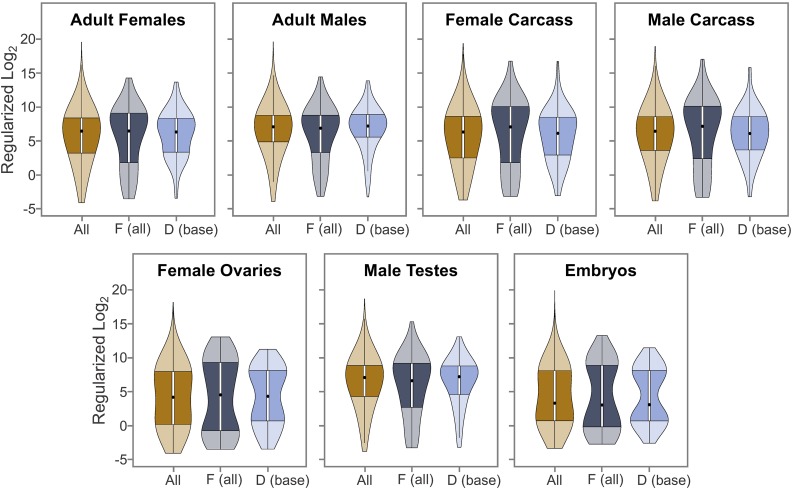
Figure 8.
D. ananassae F-element genes show similar expression patterns compared to genes on other Muller elements. RNA-Seq reads from seven samples (adult females, adult males, female carcass, male carcass, female ovaries, male testes, and embryos) were mapped against the improved D. ananassae genome assembly and the read counts for the Gnomon gene predictions were tabulated by htseq-count. The read counts for the seven samples were normalized by library size and then transformed using Tikhonov/ridge regularization in the DESeq2 package to stabilize the variances among the samples. The violin plots compare the distributions of the regularized log2 expression values for the D. ananassae Gnomon gene predictions on all scaffolds (All), on the F-element scaffolds [F (all)], and on the base of the D element [D (base)] for these different developmental stages and tissues.