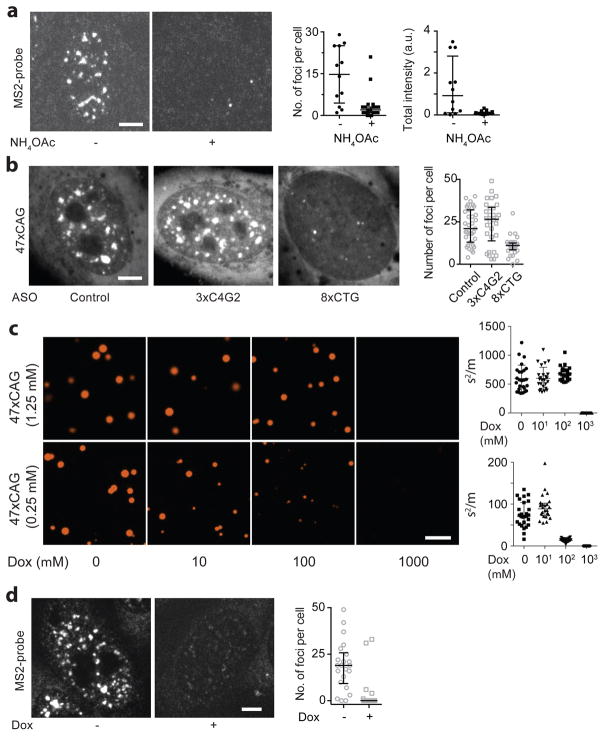
Extended Data Figure 6. RNA foci are disrupted by treatments that prevent RNA gelation in vitro.
(a) RNA FISH using a probe directed against MS2 hairpin loops confirms that 47xCAG RNA foci are disrupted by treatment with 100 mM NH4OAc, thus precluding the possibility that the observed disruption of RNA foci in live cells is due to dissociation of MS2CP-YFP from the MS2 hairpins. Representative images and corresponding quantification. (b) Transfection of an 8xCTG oligonucleotide disrupted 47xCAG RNA foci while control oligonucleotides (3xC4G2 or Control) did not. Representative images and quantification of the number of RNA foci per cell. Sequences of the oligonucleotides are provided in Supplementary Table 2. (c) Doxorubicin disrupts 47xCAG RNA clustering in vitro in a dose-dependent manner. Representative micrographs and the quantification of the inhomogeneity in the solution at indicated RNA and doxorubicin concentrations. (d) RNA FISH using a probe directed against MS2 hairpin loops confirms that 47xCAG RNA foci are disrupted by treatment with 2.5 μM doxorubicin, suggesting that the observed disruption of RNA foci in live cells is likely not an artefact of MS2CP-YFP dissociation from MS2 hairpins. Scale bars represent 5 μm. Error bars depict median and interquartile range. Data are representative of 3 or more independent experiments.