Figure 1.
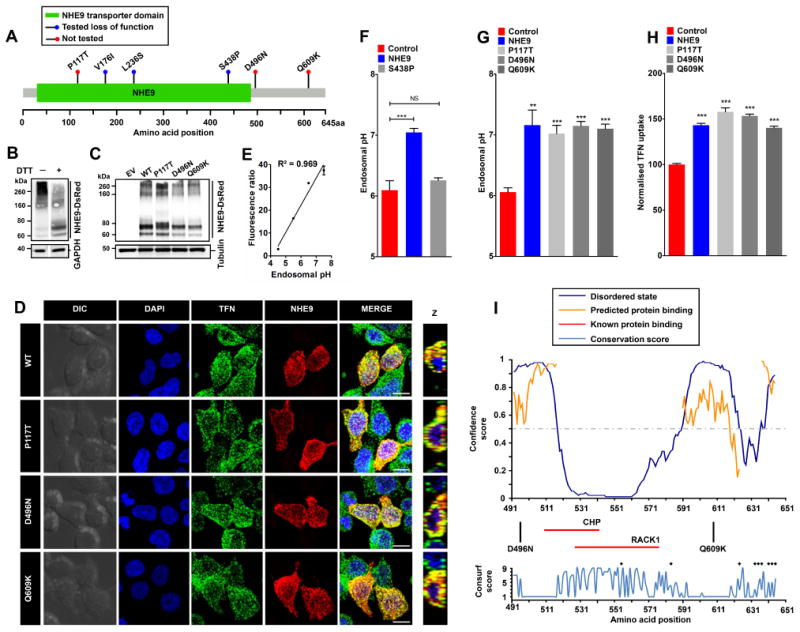
A. Gene distribution of autism associated rare missense variants in NHE9 in membrane embedded transporter domain and the C-terminal regulatory cytoplasmic domain [1] . The x-axis indicates amino acid locations of the NHE9 protein. Three of these variants (V176I, L236S and S438P) were reported as loss of function previously [2] and the remaining three (P117T, D496N, and Q609K) variants are evaluated in this study. B. Immunoblot of cell lysate from HEK293 cells expressing NHE9-DsRed using an Anti-DsRed antibody without and with dithiothreitol (DTT) treatment run on the same Western blot. Note prominent high molecular weight smear (>200 kDa) of NHE9 that was resolved into discrete band(s) with DTT treatment. C. Expression levels of NHE9 and autism-associated missense variants in HEK293 cells. Immunoblot of total lysate from cells with empty vector (EV) transfection and from cells expressing NHE9-DsRed (WT), P117T-DsRed, D496N-DsRed, and Q609K-DsRed using Anti-DsRed antibody. D. Localization of NHE9 and autism-associated variants to transferrin (TFN)-positive endosomes in HEK293 cells following 60 minutes of uptake. Confocal fluorescence images of DsRed-tagged NHE9 (WT and variants) (red) localize with Alexa Fluor 488-tagged TFN (green). Prominent colocalization can be seen in merged images and their orthogonal views (Z) by the presence of yellow puncta. Scale bar, 10 μm. E. Linear calibration curve of endosomal pH from fluorescence ratio of internalized FITC tagged TFN and Alexa Fluor 633 tagged TFN. Cells were loaded with TFN for 1 hour and then exposed to buffers with fixed pH values in the presence of 10 μM K+/H+ ionophore nigericin and 10 μM K+ ionophore valinomycin and quantified using flow cytometry (R2, square of the Pearson correlation coefficient=0.969). F. Expression of NHE9, but not the previously characterized autism-associated S438P mutation, resulted in significant alkalization of endosomal pH (mean ± S.D.; n=three biological replicates; NS, not significant; ***,P<0.001; two-tailed t test). G. Expression of NHE9 (WT and variants) resulted in significant alkalization of endosomal pH, relative to control (mean ± S.D.; n=three biological replicates; **,P<0.01; ***,P<0.001; two-tailed t test). No significant difference in endosomal pH was noted in cells expressing either WT or the three NHE9 variants. H. Expression of all three variants significantly increased steady state accumulation Alexa Fluor 633-tagged TFN to levels comparable to WT NHE9 (mean ± S.D.; n=three biological replicates; ***,P<0.001; two-tailed t test), indicating that all three autism-associated variants were not deleterious to ion transport function of NHE9. I. Upper panel shows in silico disorder (solid blue line) prediction for the NHE9 C-terminal tail. The orange line shows the confidence of disordered residues being involved in protein binding activity. The threshold above which amino acids are regarded as disordered is shown as a grey dashed horizontal line. The x-axis indicates amino acid locations of the NHE9 protein and the y-axis indicates confidence scores. The middle panel shows location of the two autism variants in the tail and binding residues for two known interacting proteins, RACK1 and CHP (horizontal red lines). The lower panel shows Consurf conservation scores for amino acids in the C-terminal tail. Intriguingly, most disordered amino acids showed poor evolutionary conservation. Black asterisks indicate amino acid residues with Consurf scores below the confidence cut-off.
