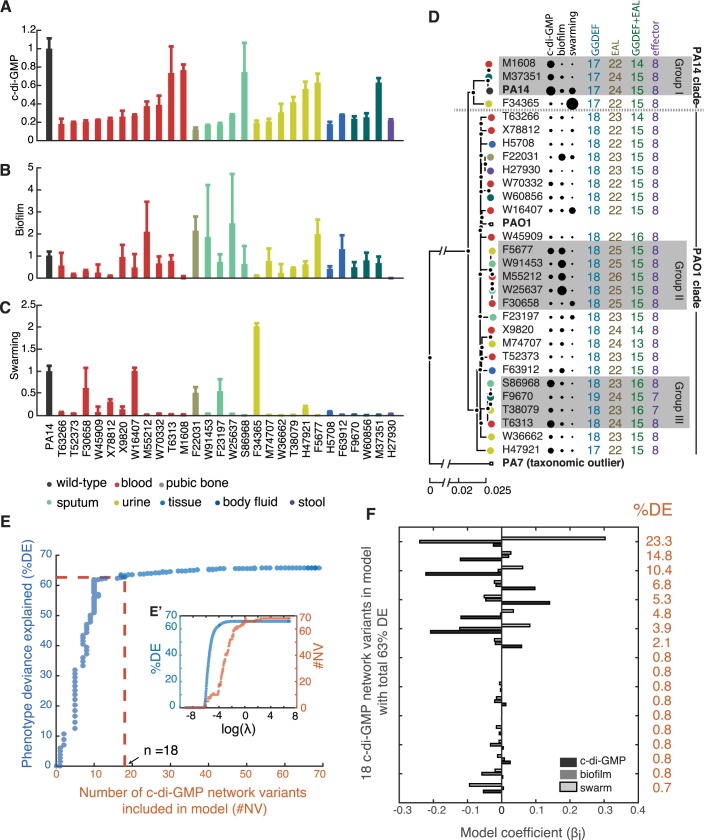
Fig 2. Phenotypic diversity in 28 P. aeruginosa isolates from acutely infected cancer patients at MSKCC explained by many small-effect alleles in c-di-GMP network.
A: Bulk c-di-GMP levels collected from bacterial colonies, including for the laboratory strain PA14. B: Biofilm levels measured in microtiter plates using the crystal-violet assay. C: Motility measured as swarm area after 16 h of incubation. D: Phylogenetic tree reconstructed from 88,347 genetic variants identified in core genes, including PA14 and two other laboratory strains PAO1 and PA7. Numbers shown represent the number of open-reading frames (ORFs) identified with c-di-GMP related motifs: GGDEF domain for synthesizing c-di-GMP, EAL for degrading c-di-GMP, and effector for sensing c-di-GMP. Some ORFs encode both GGDEF and EAL domains. E: Explaining diversity in c-di-GMP, biofilm and swarming required many alleles of small-effect in c-di-GMP genes identified within the 28 genomes. Model selection using LASSO revealed that a model that explains 85% of the phenotypic deviance requires including at least 21 genetic variants in c-di-GMP related genes. E’ shows a detail of LASSO model selection, which increases the tuning parameter λ and selects variants to include in the model. F: Each of the 21 genetic variants by itself explains 27% or less of the phenotypic variance, even in the best model selected by LASSO. The analysis supports that the phenotypic diversity observed among clinical isolates is due to small-effect alleles.