Fig. 2.
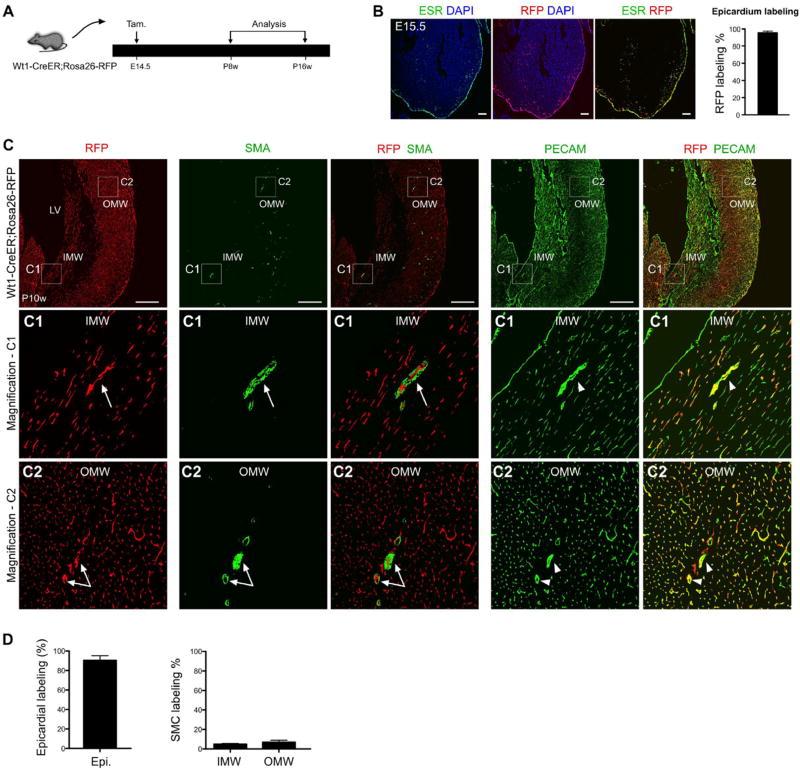
Epicardial cells labeled at E14.5 rarely contribute to SMCs of the 2nd CVP. (A) The experimental strategy for pulse-and-chase lineage tracing studies, with tamoxifen induction (the pulse) at E14.5 and analysis (the chase) from postnatal 8 weeks (P8w) to P16w in Wt1-CreER; Rosa26-RFP mice. (B) The active expression domain of WT1, as determined by ESR (estrogen receptor), and the Wt1-lineage, as determined by expression of the RFP reporter, in sections of E15.5 Wt1-CreER; Rosa26-RFP embryonic hearts. Nuclei are stained by DAPI. Tamoxifen was injected at E14.5. Labeling efficiency was calculated as the percentage of RFP + ESR + epicardial cells in total number of ESR + epicardial cells. n = 4. Scale bars, 100 µm. (C) Immunostaining of RFP, SMA, PECAM and DAPI on sections of Wt1-CreER; Rosa26-RFP hearts. Wt1-CreER labels coronary endothelial cells (RFP+; PECAM+, arrow-heads) but very few vascular SMC (RFP+; SMA+, arrows) in the IMW or OMW. Scale bars, 0.5 mm. LV, left ventricle; IMW, inner myocardial wall; OMW, outer myocardial wall. Representative figure of 4 individual samples. (D) Quantification of the percentage of epicardial cell labeling or smooth muscle cell (SMC) labeling by Wt1-CreER mediated lineage tracing (RFP+). Compared with robust epicardial cell labeling, few SMCs were labeled in the IMW and OMW. n = 4.