Fig. 3.
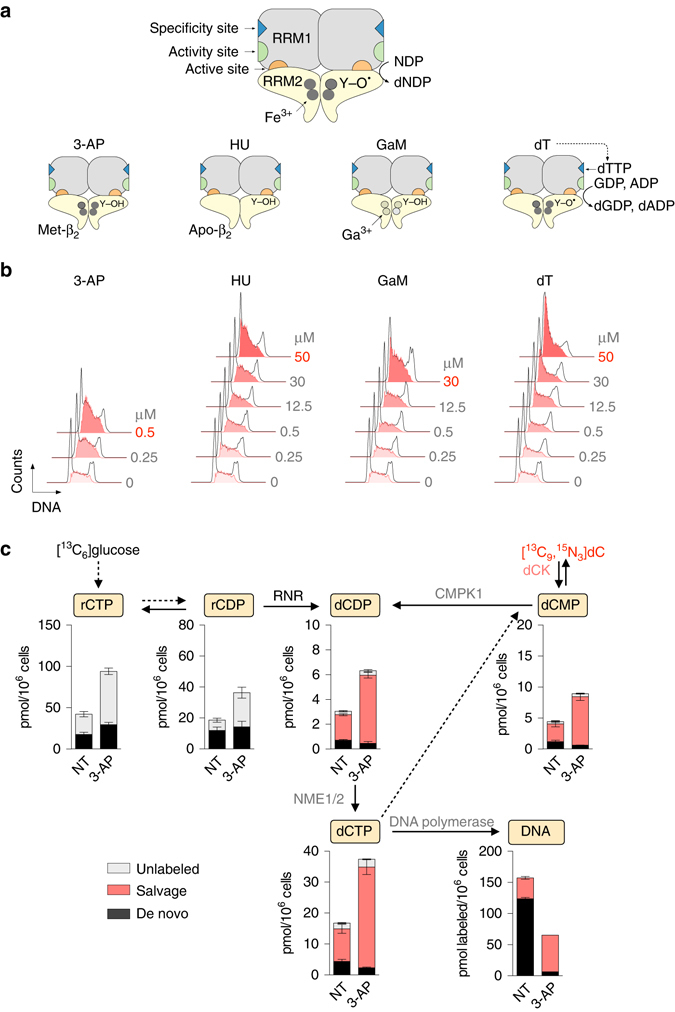
3-AP potently inhibits RNR and enhances salvage nucleotide biosynthesis. a Mechanisms of action of four RNR inhibitors. The two RNR subunits, RRM1 (α) and RRM2 (β) form a catalytically active α2β2 complex. Each RRM1 subunit contains two allosteric regulatory sites (the specificity and activity sites), as well as the active site, where nucleotide reduction occurs. The active form of the RRM2 dimer (holo-β2) houses the di-iron cofactor and the tyrosyl radical (Y-O•). 3-AP forms a complex with Fe2+ which interferes with the regeneration of the tyrosyl radical in RRM2 therefore promoting the formation of an inactive met-β small subunit which retains its di-iron center7. Hydroxyurea (HU) scavenges the RRM2 tyrosyl radical and depletes the di-iron center to form an inactive apo-β form. Gallium maltolate (GaM) releases Ga3+ which mimics Fe3+ and disrupts the RRM2 di-iron center. Thymidine (dT) is converted via the salvage pathway to thymidine triphosphate (dTTP) which binds to the allosteric specificity site on RRM1 to favor GDP reduction over pyrimidine (CDP and UDP) reduction, thereby resulting in dCTP insufficiency. b Effects of RNR inhibitors on cell cycle progression. CEM cells were incubated for 24 h with indicated concentrations of RNR inhibitors followed by cell cycle analyses using flow cytometry. Shown in bold red are the concentrations of each RNR inhibitor required to induce a greater than 45% increase in the S-phase population, indicative of S-phase arrest due to nucleotide insufficiency. Cell cycle plots are representative of two independent experiments. See Supplementary Fig. 8 for quantification. c LC-MS/MS-MRM analysis of dCTP biosynthesis in CEM cells treated with 500 nM 3-AP for 12 h (mean ± s.d., n = 3). NT not treated. CMPK1 uridine-cytidine monophosphate kinase 1, NME1/2 nucleoside diphosphate kinase 1/2, dC 2’-deoxycytidine, dCMP deoxycytidine monophosphate, dCDP deoxycytidine diphosphate
