Figure 9. KPAF3 recognizes G‐rich sequences characteristic of pre‐edited mRNAs.
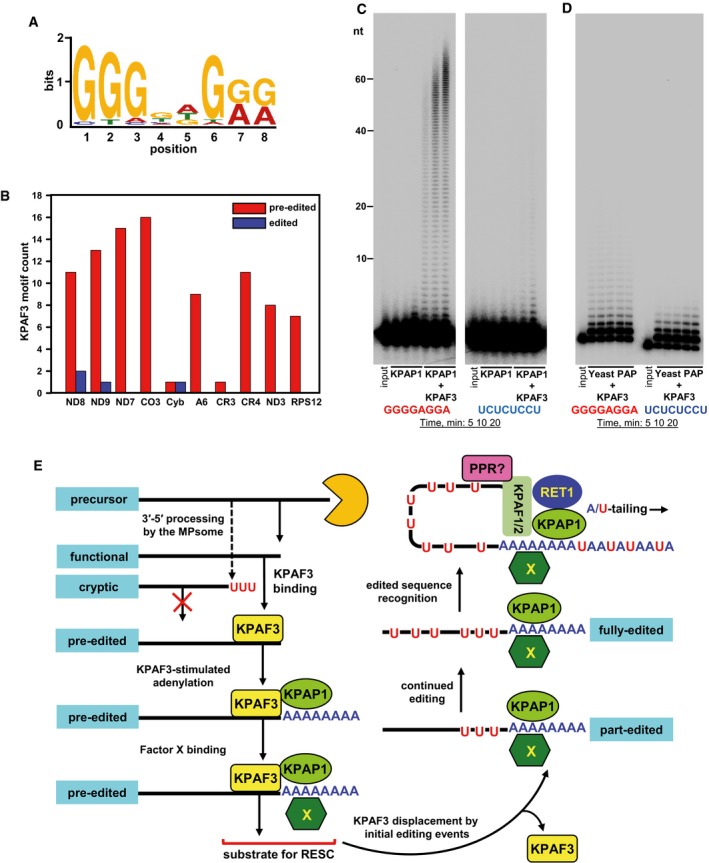
- KPAF3 in vivo binding motif. The MACS algorithm was used to call KPAF3 CLAP‐Seq peaks separately on both maxicircle strands. The significant peaks from samples treated with low and high RNase I concentrations were extended on both sides by 100 nt and used as input; the maxicircle sequences were used as the background model. The MEME algorithm was applied to predict the enriched motif for KPAF3 binding.
- Distribution of KPAF3 binding sites between pre‐edited and edited transcripts. The motif shown in panel (A) was queried against edited and pre‐edited maxicircle transcripts using FIMO algorithm with a P‐value cutoff at 0.001. The number of predicted motifs in pre‐edited and edited maxicircle transcripts was plotted as a bar graph.
- Motif‐dependent stimulation of KPAP1 poly(A) polymerase by KPAF3. Synthetic 5′‐radiolabeled 40‐mers containing either predicted G‐rich motif (left panel), or arbitrary pyrimidine octamer (right panel), was incubated with 100 nM of KPAP1 in the presence or absence of 100 nM of KPAF3. Reactions were performed for 5, 10, 20 min, and products were resolved on 10% polyacrylamide/8 M urea gel.
- Specificity of KPAP1 poly(A) polymerase stimulation by KPAF3. The assay was performed with 14 nM of yeast poly(A) polymerase in the presence of 0, 25, 50, 100, and 200 nM of KPAF3 for 20 min, and products were resolved on 10% polyacrylamide/8 M urea gel.
- Model for functional coupling of primary precursor processing, adenylation, and editing processes. The MPsome‐catalyzed 3′–5′ degradation pauses near the mature 3′ end by a still‐unknown mechanism. Upon pausing, however, two outcomes become feasible depending on the KPAF3 binding site's proximity to the 3′ end: (i) KPAF3 recruits KPAP1 poly(A) polymerase and stimulates short A‐tail addition to downstream terminus; and (ii) lack of bound KPAF3 causes MPsome to dissociate leaving either the unmodified 3′ end, or that with RET1‐added U‐tail. The former modification likely designates the transcript as mRNA, while the latter occurs on rRNAs and truncated mRNA species. A hypothetical factor X is proposed to bind the A‐tail to stabilize edited mRNA once the editing machinery displaces KPAF3 from the 3′ region. Addition of long A/U‐tail to a pre‐existing 3′ A‐tail is triggered upon completion of editing, which typically occurs at the 5′ end. Hence, we hypothesize the existence of a PPR factor that recognizes the RNA sequence created de novo by editing, and recruits KPAF1/2 factors and RET1 TUTase to short A‐tail preloaded with KPAP1 and Factor X. This event likely triggers A/U‐tailing, leading to translational activation (Aphasizheva et al, 2011).
