Figure 1. Genome-wide Association Study of Survival in Patients With Sporadic Amyotrophic Lateral Sclerosis (ALS).
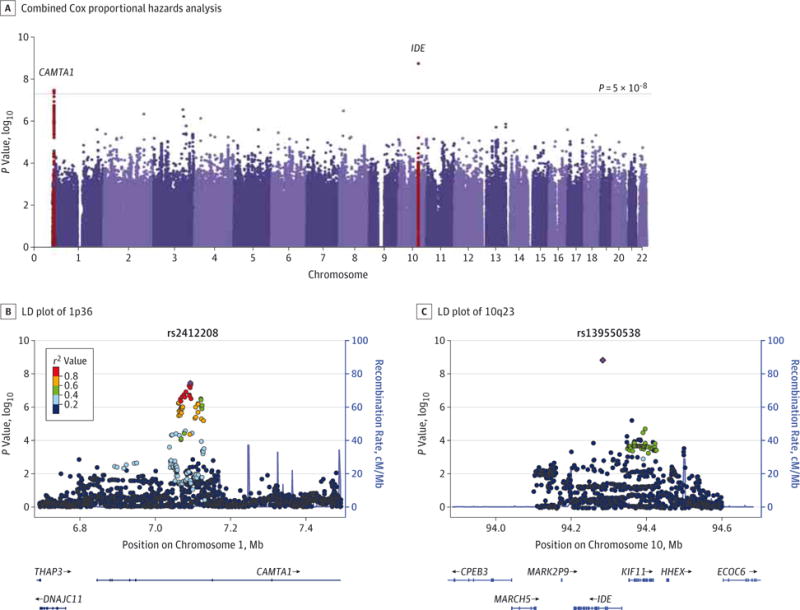
A, Manhattan plot of the combined (METAL software) Cox proportional hazards regression analysis. The threshold for genome-wide significance after correction for multiple testing was set at P = 5 × 10−8 (horizontal blue line). Loci significantly associated with ALS survival are highlighted in red and labeled according to the corresponding genes. At locus 10q23, the most associated single-nucleotide polymorphism (SNP), rs139550538 (P = 1.87 × 10−9), was moderately rare with a minor allele frequency (MAF) of 0.03, whereas at the 1p36 locus, the 4 SNPs significantly associated (rs2412208 [P = 3.53 × 10−8], rs4584415 [P = 3.68 × 10−8], rs35447019 [P = 3.86 × 10−8], and rs4409676 [P = 4.48 × 10−8]) were common (MAF > 0.26). B and C, Regional linkage disequilibrium (LD) plots of the 2 regions significantly associated with ALS survival. At the 1p36 locus, 4 SNPs passed the genome-wide significant threshold, followed by 87 tagged proxies suggestively associated (P < 10−4). All the associated SNPs mapped within introns 3 to 4 of the CAMTA1 gene. At the 10q23 locus, the top-ranked SNP, rs139550538, intronic to the IDE gene, was in weak (r2 < 0.4) LD with the tagged proxies that were located in the neighbor gene, KIF11.
