Fig. 5.
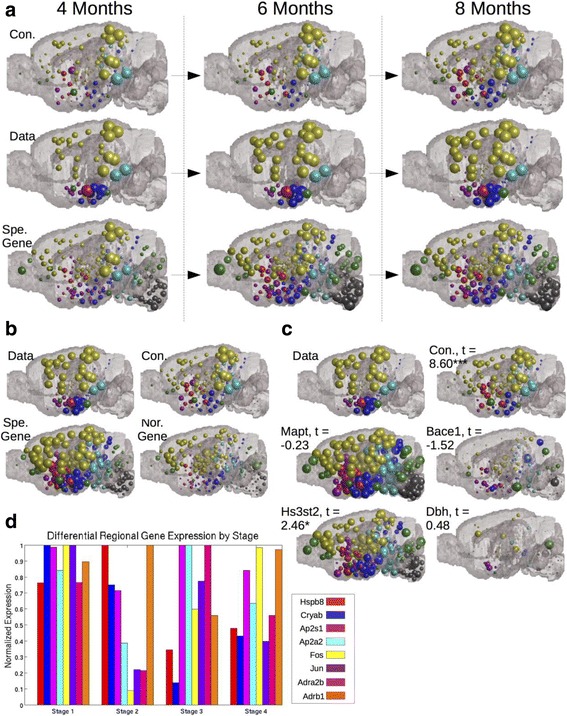
ND modeling indicates connectivity is a better predictor of tau pathology progression and regional vulnerability than regional gene expression but that regional gene expression does better in the non-seeded mouse dataset than in seeded datasets. a An anatomic spatiotemporal illustration of the predictions of ND modeling using both the anatomic connectivity network and the gene expression similarity network, across a specific subset of genes known to be important for promoting tau aggregation and transcription, as compared with the empirical spatiotemporal data on tau progression patterns. b An anatomic illustration of the results from Tables 2 and 3—bottom-section comparing ND using the connectivity network with absolute regional gene expression (c) Modeling pathology using individual genes known to have a mechanistic link with proteinopathy (Mapt, Bace1, Hs3st2) or genes necessary for noradrenergic neurotransmission (Dbh) does not perform as well as ND using the anatomic connectivity network when analyzed using all 426 ABA regions. d Genes that are differentially expressed among regions showing baseline tau relative to the rest of the brain are not more heavily expressed in regions showing pathology at baseline or early relative to later stages. The color and location legends for the major regions in the brain illustrations are the same as in Fig. 2 and Fig. 4, and sphere size corresponds with degree of predicted pathology in a given area. * p < 0.05, ** p < 0.01, *** p < 0.001
