Abstract
Introduction
Natural products from culture collections have enormous impact in advancing discovery programs for metabolites of biotechnological importance. These discovery efforts rely on the metabolomic characterization of strain collections.
Objective
Many emerging approaches compare metabolomic profiles of such collections, but few enable the analysis and prioritization of thousands of samples from diverse organisms while delivering chemistry specific read outs.
Method
In this work we utilize untargeted LC–MS/MS based metabolomics together with molecular networking to
Result
This approach annotated 76 molecular families (a spectral match rate of 28 %), including clinically and biotechnologically important molecules such as valinomycin, actinomycin D, and desferrioxamine E. Targeting a molecular family produced primarily by one microorganism led to the isolation and structure elucidation of two new molecules designated maridric acids A and B.
Conclusion
Molecular networking guided exploration of large culture collections allows for rapid dereplication of know molecules and can highlight producers of uniques metabolites. These methods, together with large culture collections and growing databases, allow for data driven strain prioritization with a focus on novel chemistries.
Keywords: Metabolomics, Marine natural products, Mass spectrometry, Molecular networking
1 Introduction
Since Alexander Fleming’s (1929) discovery of penicillin from a cultured microbe and its subsequent utilization as a potent anti-infective, many biotechnology and biomedicine sectors have come to rely on culture collections for the discovery of new molecules (Fleming 1929; Smith 2003). Culture collections provide an invaluable view into the world of over 1030 prokaryotes on this planet that are suspected to contain half of all biologically incorporated carbon (Whitman et al. 1998). While only a small fraction of these organisms can be grown in the lab, investigations of these collections have yielded many new chemistries and provided a driving force for clinical and biotechnological progress. Microbes have evolved to survive and thrive in every conceivable niche on this planet. One adaptation that has facilitated this diversification is the production of specialized chemistries. Indeed, the specialization of both primary metabolism, but in particular secondary or specialized metabolism can lead to the production of diverse natural products that are functionally optimized for the producer’s niche (Kinkel et al. 2014). Both biotic features (neighboring micro- and macro-organisms) as well as the abiotic environment, such as the nutrient content of terrestrial and marine sediments, determine the ability of microbial communities to co-exist and thrive (Quinn and Alexandrov 2014; Kelly et al. 2014). As such, obtaining community members in culture provides invaluable opportunities to explore the diverse chemistries produced in nature.
Metabolomics, particularly mass spectrometry (MS) based metabolomics, is ideally suited for taking an inventory of the chemistry associated with culture collections (Dunn et al. 2012; Kersten et al. 2011). Many groups have recognized that liquid chromatography (LC) enhanced MS metabolomics is a powerful strategy to peer into the diversity of natural products from culture collections and as such LC–MS based metabolomics has been used frequently in natural product discovery programs. The goal of such metabolomics based efforts has been to identify both known (i.e. dereplicate) and unknown molecules in complex backgrounds. The most common approaches utilized in natural product dereplication and discovery programs are MS1 based or UV–vis based spectral matching (Nielsen et al. 2011). No large public database exists for this information and thus these approaches are limited to in-house data. Multivariate statistics such as principle component analysis (PCA) has been applied to such data for prioritizing strains from small collections and has the capability to identify unique molecules (Larsson et al. 2007; Hou et al. 2012). However, large scale MS/MS, also described as MS2 or tandem mass spectrometry, based approaches are only beginning to be utilized in similar natural product discovery programs. While MS1 based approaches are useful for dereplicating known molecules and identifying potential discovery targets in a small number of samples, tools such as PCA become less effective when the numbers of samples reach into the hundreds (Kellogg et al. 2016). For example, the existing workflows are unable to assess the frequency of chemistries associated with a given culture collection. Prior natural product metabolomics workflows also cannot address the following questions: How many known molecules are there? How many related molecules can be detected in a culture collection? What are the unique molecules in such collections? What extracts contain the largest number of unique molecules? Although there has been a surge in the development of exciting and useful metabolomics analysis tools and resources over the past few years, none can address the above questions (Tautenhahn et al. 2012). In addition, the majority of analysis resources that are available tend to be designed to handle the annotation of individual spectra, primary metabolism of human or model systems, or the simultaneous comparison of related cohort data sets (Wishart et al. 2013; Sawada et al. 2012; Guo et al. 2013). Most metabolomic tools are not designed to extract information from datasets obtained from very different samples as would be expected from diverse culture collections.
In response to such needs, the Global Natural Product Social (GNPS) molecular networking platform was recently launched (Wang et al., accepted). It is a crowd sourced knowledge repository and analysis infrastructure launched to capture mass spectrometry expertise from the community and to begin addressing such aforementioned questions using the analysis based on molecular networking. Molecular networking groups MS/MS spectra based on spectral similarity. One can include reference MS/MS spectra from trusted standards or community annotations to dereplicate known molecules as a level two, putatively annotated, or level 3, putatively characterized compound as defined by the metabolomics initiative (Sumner et al. 2007). Such spectral comparisons are displayed as a graph or network, resulting in an information-rich map of the mass spectrometry detectable chemical space. This type of analysis, since its introduction in 2012, is becoming widely adopted by the natural product and metabolomics community. For example, it has been adapted to more rapidly elucidate the structure of a human-associated toxin (Vizcaino et al. 2015), optimize in silico fragmentation libraries (Allard et al. 2016), build activity guided discovery programs (Kurita et al. 2015), and to enhance and automate SILAC studies (Klitgaard et al. 2015). Here we use molecular networking to display and navigate the mass spectrometry detectable chemical space of 3000 microbial extracts derived from 1000 distinct marine organisms.
Roughly half of all prokaryotes can be found in the marine environment (Kallmeyer et al. 2012). The Scripps Institution of Oceanography (SIO) houses large collections of these marine-derived microbes isolated from many different locations around the world. This collection includes a large number of actinomycetes, members of a bacterial order responsible for many molecules useful to biotechnologists and clinicians (Bérdy and View 2005; Subramani and Aalbersberg 2012). Among the many molecules discovered from this class of microorganisms are the antibiotics vancomycin and daptomycin (Baltz et al. 2005), the immunosuppressant rapamycin (Sehgal et al. 1975), and avermectin (Burg et al. 1979), the latter of which resulted in the 2015 Nobel prize in medicine for the treatment of worm infections. Over twenty thousand molecules have been discovered from marine sources (Gerwick and Moore 2012). Several thousand of these were shown to be bioactive, with half of that activity observed against cancer cell lines (Hu et al. 2015). Over the years, marine natural products have led to the assessment of more than a dozen molecules in clinical trials and several marine natural products are approved by the FDA (Gerwick and Moore 2012). In addition to anti-tumor and anti-infectives, some marine derived peptides have shown promise in the treatment of Alzheimer’s disease and other indications (Russo et al. 2015). Culture collections at SIO have likewise been a productive source of novel natural product derived therapeutics (Fenical and Jensen 2006) including salinosporamide A (Fenical et al. 2009), which is in clinical trials for the treatment of cancer as well as the anti-inflammatory skin treatment, honaucin A (Choi et al. 2012; Anon n.d.).
In the last century, much effort revolved around the discovery of chemistry from terrestrial sources. However, marine microbes are, comparatively, an understudied source of chemistries and yet remarkably productive from a biotechnology standpoint (Montaser and Luesch 2011; Blunt et al. 2014). We therefore set out to assess the discovery potential of 1000 randomly selected microorganisms through untargeted MS-based metabolomics analysis of a high throughput culturing effort using an SIO marine microorganism collection. Using a combination of traditional multivariate statistics and molecular networking, we are able to characterize the contents of a large diversity of chemical space, as well as to locate novel metabolites within the sampled space.
2 Materials and methods
2.1 High throughput culturing
2.1.1 Microorganism selection
To enable metabolomics analysis, one thousand marine isolates were selected from a culture collection maintained at the Scripps Institution of Oceanography. Roughly 20 % of strains were specifically selected due to previous investigation and known production of interesting natural products, while the rest were randomly selected out of the collections database. This cohort represents a diverse collection of strains with the bulk (over 70 %) being taxonomically uncharacterized.
2.1.2 Growth and extraction
The 1000 microorganisms were cultured from frozen stocks in 96-well format. Three extracts were analyzed using two culture growths. Ten μL of each frozen stock was inoculated into 500 μL of Salt Water ISP2 media (0.4 % Yeast Extract, 1 % Malt Extract, 0.4 % Dextrose, 2.2 % InstantOcean salts (Instant Ocean, Spectrum Brands, Backburg, VA, USA) (w/v)) in deep welled polypropylene 96-well plates (Thermo Fisher Scientific, Waltham, MA, USA). These starter cultures were covered with breathable sterile seals (AeraSeal (Excel Scientific, Victorville, CA, USA) and incubated for 7 days at 28 °C while shaking at 200 RPM. A 5 μL aliquot of each starter culture was then inoculated into deep-well 96-well plates containing 500 μL of solid media (Salt Water ISP2 supplemented with 2 % agar) and grown for 7 days at 28 °C without agitation. After inoculation, the remainder of the starter plates were brought to 20 % glycerol and stored at −20 °C. The cultures were extracted with acetonitrile and water (1:1 v/v) by adding 500 μL of the extraction solvent to the colony surface, sonicating at 40 kHz for 10 min in a Branson 5510 ultrasonic water bath (Branson Ultrasonics, Danbury, CT, USA), and transferring the top 200 μL of solvent and to new deep welled plates. A second set of cultures were generated in a similar manner by inoculating the agar surface directly from the frozen stocks. These plates were cultured in the same manner and sequentially extracted with 1:1 ethyl acetate:methanol followed by butanol. For all extracts cell debris was removed by centrifugation at 2500×g and the top 100 μL of extract transferred to clean 0.5 mL 96-well polypropylene plates (Agilent Technologies Inc., Santa Clara, CA, USA), dried in air at room temperature, and dissolved in 100 μL of methanol containing 10 μM glycocholic acid, a compound not expected to be produced by the marine organisms, as an internal standard to monitor chromatographic behavior during LC–MS and to ensure equal injection volumes during LC–MS/MS analysis.
2.1.3 Untargeted metabolomic profiling by UPLC-ESI-Q-TOF–MS/MS
Null injection and solvent only blanks were acquired before each 96-well plate of extracts and media controls were acquired after washing. Twenty microliters of sample were subjected to chromatographic separation with an Agilent 1290 Infinity UPLC equipped with a Kinetex C18 column (1.7 μm, 50 × 2.1 mm) (Phenomenex, Torrence, CA, USA) held at 30 °C. A ten minute mobile phase gradient (0 min: 5 % B, 1 min: 5 % B, 3 min: 80 % B, 5 min: 100 % B. 8 min: 100 % B, 9 min: 5 % B, 10 min: 5 % B) where solvent A is 98 %H20, 2 %ACN, 0.1 % Formic Acid and solvent B 98 %ACN, 2 %H20, 0.1 % Formic Acid. Column eluent was analyzed using a Bruker Micro-TOF-QII mass spectrometer equipped with an ESI source operating in positive polarity. The UPLC system and mass spectrometer were controlled by otofControl and Hystar software packages (Bruker Daltronics Inc., Billerica, MA, USA). Data, MS1 and MS2, were acquired in a data dependent manner at 3 Hz, fragmenting the ten most abundant precursor ions per MS1 scan, acquiring MS/MS data between 200 and 2000 m/z. The instrument was externally calibrated once every 24 h using ESI-L Low Concentration Tuning Mix (Agilent Technologies). Hexakis (1H,1H,3H-tetrafluoropropoxy)phosphazene (Synquest Laboratories Inc., Alachua, FL, USA), m/z 922.009798, was introduced as an internal calibrant (lock mass) during the all runs. Instrument parameters were set as follows: nebulizer gas (nitrogen) pressure, 3 Bar; dry gas flow, 9 L/min; Capillary voltage, 4500 V; ion source temperature, 200 °C; spectra rate acquisition, 3 spectra/s. MS/MS fragmentation of the ten most intense selected ions per spectrum was performed using ramped collision-induced dissociation energy of 16–112 eV, applied based on parent mass. Repetitive MS/MS sampling was limited by exclusion after 3 spectra at a particular mass were acquired. This was released after 30 s. A permanent MS/MS exclusion list criterion was set to prevent oversampling of the internal calibrant.
2.2 MS/MS data processing and analyses
2.2.1 Molecular Networking
Raw Bruker analysis files had lock mass calibration applied and were converted to mzXML format with Bruker’s Data Analysis software and uploaded to the global natural product social (GNPS) molecular networking tools. The data itself is accessible in GNPS with accession numbers MSV000078787, MSV000078936, and MSV000078937. In GNPS we subjected the data to molecular networking. The resulting analysis and parameters for the network can be accessed via this link http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=02c9beb84d804ff180af5554f500bac8. The details of the settings are described below as a part of the verbatim description that is automatically generated on GNPS to describe this network. A molecular network was created using the online workflow at GNPS. The data was filtered by removing all MS/MS peaks within ±17 Da of the precursor m/z. The data was then clustered with MS-Cluster with a parent mass tolerance of 2.0 Da and a MS/MS fragment ion tolerance of 0.5 Da to create consensus spectra. Further, consensus spectra that contained less than 2 spectra were discarded. A network was then created where edges were filtered to have a cosine score above 0.7 and more than five matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top ten most similar nodes. The spectra in the network were then searched against GNPS’s spectral libraries. All matches between network spectra and library spectra were required to have a score above 0.6 and at least 6 matched peaks. “Some analysis was carried out through the GNPS web interface, while more advanced network analysis was exported from GNPS and then analysis conducted in Cytoscape per instructions in the documentation of GNPS, https://bix-lab.ucsd.edu/display/Public/GNPS+Documentation+Page (Shannon et al. 2003).
2.2.2 Multivariate analyses
The results of GNPS were exported as a matrix of identical spectra, or nodes, observed in each sample. These data were used to construct a sample to sample distance matrix calculated using the well known binary Jaccard dissimilarity index (1912). Subsequent principal coordinate analysis (PCoA) dimensional reduction was visualized using the Emperor visualization platform (Vázquez-Baeza et al. 2013).
2.3 Novel metabolite characterization
2.3.1 Isolation of maridric acids A and B
Strain CNP-993 was grown on salt water ISP2 media (yeast extract 0.4, malt extract 1.0 %, glucose 0.4 %, and instant ocean 2.2 %, pH 7.3). The whole broth (1 L) fermented for 7 days at 28 °C was extracted with ethyl acetate. The ethyl acetate layer was separated from the aqueous phase, filtered, and concentrated in vacuo to dryness. The crude extract (251 mg) was solubilized in a small amount of chloroform, and applied on a silica gel column (5.7 g). The material was eluted stepwise with the mixtures of chloroform and methanol (100:0, 100:1, 50:1, 20:1, 5:1 and 1:1, each 40 mL). The fractions were analyzed by LC–MS using an HPLC1260 system (Agilent) equipped with a Kinetex C18 column (Phenomenex, 5 μm, 4.6 × 150 mm2). A 20-min linear gradient from 70 to 100 % acetonitrile in water containing 0.1 % formic acid was employed at a flow rate of 1 mL/min. Eluent was analyzed by an Amazon ETD (Bruker) ion-trap mass spectrometer equipped with an ESI source and operated in positive polarity, and CID for MS/MS fragmentation. The 20:1 fraction that contained the targeted ions with parent masses of 971.62 and 1359.87 m/z was concentrated in vacuo to dryness to give a yellow material (52.1 mg). This material (8.0 mg) was finally purified with LC–MS (column; SUPELCO C18 (SIGMA-ALDRICH, 5 μm, 10 × 250 mm2) (SIGMA-ALDRICH, St. Louis, MO, USA), solvent; 95 % CH3CN containing 0.1 % formic acid, flow rate: 2 mL/min, other conditions; the same as the above). The fractions were concentrated under reduced pressure and lyophilized to dryness to yield maridric acid A (2.0 mg) and maridric acid B (1.0 mg) as colorless oils.
2.3.2 Structure elucidation of maridric acids A and B
For NMR data acquisition approximately 1 mg of purified sample was dissolved in 50 μL of CDCl3 (Cambridge Isotope Lab, Tewksbury, MA, USA). All NMR experiments were performed on a Bruker Advance III 600 MHz spectrometer at 298 K with a 1.7 mm Micro-CryoProbe. The pulse sequences used were those supplied by Bruker, and processing was done with the Bruker TOPSPIN software. Proton, DQF-COSY, 1H–13C HSQC and 1H–13C HMBC spectra were measured. The 1H–13C HSQC spectra were recorded with delays corresponding to 1 J H–C coupling constants of 140 Hz. The 1H–13C HMBC spectra were recorded with delays corresponding to 2 or 3 J H–C coupling constants of 8 Hz and 1 J H–C coupling constants of 145 Hz. Additional fragmentation MS experiments were performed on a Thermo LTQ-FT mass spectrometer (Thermo Electron Corp.). Aliquots from solutions of purified maridric acids A and B were diluted in acetonitrile.
2.3.3 MS analysis of an acetylated derivative of the maridric acid A
In order to determine the number of hydroxyl moieties in each of the subunits of the polymeric compounds, it was necessary to derivatize each moiety, observing the corresponding mass shift by tandem MS. Thus the hydroxyl moieties of maridric acid A (100 μg) were selectively acetylated by treatment with acetic anhydride and pyridine (each 100 μL) (Chen et al. 1996). After 16 h, at room temperature, the mixture was diluted with 1.0 N HCl, and the aqueous phase was extracted with ethyl acetate. The organic layer was recovered and concentrated in vacuo to dryness. Then, a part of the sample was analyzed by LC–MS using an HPLC1260 system equipped with a XBridge column (Waters, 5 μm, 4.6 × 150 mm2) (Waters, Milford, MA, USA) under the condition of an isocratic mobile phase of 95 % acetonitrile in water (JT Baker, Avantor Performance Materials, Center Valley, PA, USA) containing 0.1 % formic acid (Optima, Thermo Fisher Scientific) at a flow rate of 1 mL/min. MS/MS spectra of the sample was obtained using an Amazon ion-trap mass spectrometer with nominal mass accuracy (Bruker).
3 Results and discussion
To assess the discovery potential across 1000 marine microbes we endeavored to exhaustively sample the MS-visible chemical space produced by these organisms under standard laboratory conditions. To this end we developed a high throughput pipeline for small scale culturing and extraction. Cultures were extracted with three different solvent systems and subjected to data dependent UPLC-MS/MS analysis. By utilizing multiple solvent systems, we were able to extend the chemical space covered and access slightly different arenas of polarity. By using untargeted data acquisition, it was possible to capture an unbiased inventory of the most abundant chemistries present in the extracts of each culture. This inventory consisted of the MS/MS fragmentation patterns of hundreds of thousands of different MS features. Manual curation of data sets of this size can be unfeasible, especially for smaller teams or those without extensive mass spectrometry expertise. In order for an MS based pipeline to be broadly applicable and rapid it was necessary to process the MS/MS data automatically and use higher-level visualizations to guide the investigations.
In order to simplify the 3000 MS/MS datasets, we analyzed the data using multivariate statistics and the global natural products social molecular networking platform (http://gnps.ucsd.edu) (Wang et al., accepted), which contains multiple open access MS-analysis tools as well as libraries of reference MS/MS spectra. Primary among these tools is molecular networking, which provides insight into the chemical similarity of all metabolites observed by MS/MS (Watrous et al. 2012). The first step in the molecular networking pipeline is the consolidation of highly similar spectra using MS-cluster, which utilizes dot-product or cosine metric with adaptive thresholds based on input data to control false clustering rates (Frank et al. 2008). Clustering reduces the number of total spectra to be compared, while maintaining the number of unique spectra. This analysis yields a list of unique metabolites observed by MS/MS analysis in each sample.
Because multivariate statistics provide a chemical overview of the individual samples, we performed PCoA using a binary Jaccard dissimilary index of the merged GNPS features from all three datasets. The PCoA dimensional reduction was visualized using the Emperor visualization platform (Vázquez-Baeza et al. 2013). This analysis (Fig. 1a) informed us that the data grouped primarily by extraction method suggesting that different compounds are accessed by each solvent system. The fact that so much of the difference between each sample depends on solvent rather than the producer’s identity may indicate that a large core of solubilized metabolites is broadly shared and distinctions between specific organisms occur on a finer scale in the metabolome. It is clear that inside the solvent driven differences in chemistry there are strain specific chemistries causing additional separation. However, this style of multivariate analysis does not provide direct insight into how much of the chemistry is different nor can we use such multivariable statistics for strain prioritization. Strain prioritization based on multivariable statistics such as PCA, which performs dimensional reduction directly on metabolite levels, has worked well for other laboratories, but appears to work best when the number of analytes are small compared to the number of organisms and samples compared in this study (Kim et al. 2009).
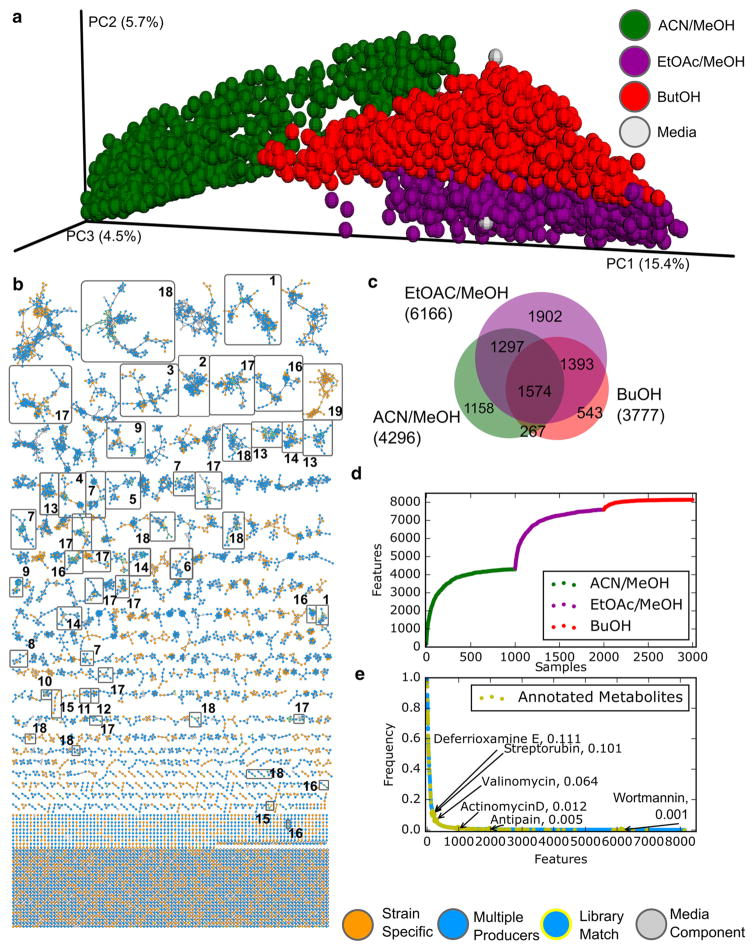
Fig. 1.
Here we show global relationships among samples through three extractions Acetonitrile/Methanol, ethyl acetate/methanol, and butanol (green, purple, and red, respectively) in PCoA space. a Media controls in grey cluster together with their corresponding extracts. The compound level inventory is displayed in the molecular network b with families containing compounds of interest boxed and the nodes with library annotations highlighted in yellow. The most notable include: valinomycin (1), nonactins (2), glucopericidin (3), actinomycin D (4), alteramide A and B (5), lobophorin (6), acyl and amphipathic (des)ferrioxamines (7), echinomycin (8), streptorubin and undecylprodiginine (9), piericidin A1 (10), antipain (11), stendomycin (12), surfactins (13), nodularin (14), wortmannin (15), ethanolaomines (16), various lipid families (17), formate clusters (18) and maridric acids (19). Chemistries observed in a single microorganism are orange, while those observed in multiple microorganisms are blue. Molecular overlap of each extraction method is displayed in a proportional Euler diagram (c). The merged rarefaction curves (d) of each extraction method shows the observed chemistries gained by additional sampling from each extraction, while the frequency analysis (e) of each metabolite shows that there is a large low abundance diversity throughout the sample set and that spectral networking is able to putatively identify metabolites at all levels of abundance (Color figure online)
We therefore subjected the data to molecular networking, yielding a metabolite-level view of the data. After unique clusters of spectra were merged with MS-cluster, only those spectra with two or more spectra per cluster in the 3000 data sets were considered for further evaluation. This removes many noisy and chimeric spectra from the analysis, as their spectral fingerprints should not be reproduced. In total, 302,847 spectra were passed through MS-cluster filters and merged to make 8155 nodes, representing unique spectra. These nodes, ranging from 190 to 1826 m/z, were then subjected to molecular networking and dereplication (Figure S1a). The network was exported from GNPS and visualized with Cytoscape (Shannon et al. 2003). This visualization (Fig. 1b) is descriptive of the entire MS-visible chemical space from these thousand isolates while still displaying information about individual metabolites. Each node represents a single chemical entity present in one or more extract. Groups of connected nodes describe molecular families, whose parent masses may differ but whose MS/MS spectra are more similar to each other than to any other merged spectrum in the sample set (Nguyen et al. 2013).
Color-coding the network based on extraction solvent revealed molecular level trends that confirm what was seen in the sample-level multivariate analyses. Determining which solvent systems extracted each unique feature (Fig. 1c) shows that ethyl acetate/methanol (1:1) appears to capture the most unique molecules, while acetonitrile/methanol (1:1) provided the second most unique molecules followed by butanol. Likewise, the accumulation of new chemistries (using unique MS/MS spectra as the definition for detecting additional chemistries) with each additional sampling event can be shown as a rarefaction curve (Fig. 1d). These merged rarefaction curves indicate that within each extraction we are approaching saturation of the observable chemical information within the 1000 strains we are investigating but that additional solvent systems provide more observable chemical diversity. The most direct comparison can be drawn between the extractions with ethyl acetate–methanol and acetonitrile-methanol, of which the former contributes most to the overall data set (Figure S1b).
During network generation, consensus spectra were also compared in the same manner to spectra from the libraries available through GNPS. Currently, over two hundred thousand spectra are available on GNPS. This includes over ten thousand in-house spectra generated and curated by the global GNPS community, as well as access to spectral databases like MassBank Japan, Europe and North America, HMDB, ReSpect and we searched the data against NIST 2014. (Horai et al. 2010; Sawada et al. 2012) The dereplication parameters used here are expected to yield low rates of incorrect spectral matching (Wang et al., accepted). In total, 290 of 8145 nodes were dereplicated and are contributed by 85,375 spectra that matched to reference spectra of known molecules. This is a match rate of 3.5 % (based on nodes) and 28 % (based on spectra). These are “level two” annotations according to the metabolomics standards defined in 2007 (Sumner et al. 2007). Due to the similarity driven organization of a molecular network, annotation of any individual chemistry also provides information about the rest of the molecular family (Yang et al. 2013). Taking this into account, molecular networking is able to provide chemical insight for 76 of 919 molecular families within this network representing nearly 19 % of all observed nodes and over 44 % of all spectra (Fig. 1b).
One of the largest and most widely distributed molecular families contained the potent potassium ionophoric depsipeptide, valinomycin (1), which was observed in over 10 % of strains (Marrone and Merz 1995). Next, the family of the nonactins (2) were dereplicated and found to contain multiple known acylated analogs identified by parent ion mass differences. However, one nonactin analog appears to be a variant not reported before, containing an additional methyl mass shift (Figure S2). Several additional families containing notable natural products were likewise dereplicated (Fig. 1b), including the following known actinomycete specialized metabolites: glucopericidin (present in 1.6 % of strains) (3), actinomycin D (3.2 %) (4), alteramide B (7.7 %) (5), lobophorin (6), acyl-desferrioxamines and amphipathic ferrioxamines (12.2 %, 11.3 %, respectively) (7), echinomycin (5 %) (8), streptorubin and undecylprodiginine (12 %, 15.8 %) (9), piericidin A1 (2.3 %) (10), antipain (0.5 %) (11), and stendomycin (\1 %) (12). Dereplication also revealed the presence compounds previously reported as Bacillus, cyanobacterial, and fungal metabolites such as the surfactins (13), nodularin (14), and wortmannin (15) highlighting the broad range of chemistries observed during this survey. Besides these families of natural products, we observed lipids (16, 17) as well as solvent derived background signals such as sodium formate clusters (18) that were also assigned by GNPS. The average metabolite was observed in 1.4 % of the strains with some appearing ubiquitously, while others were unique to single organisms or extracts.
In addition to matching against real instrument data, GNPS has begun including in silico spectral NRPS and RIPP prediction as a new method for peptide natural product dereplication (Mohimani and Pevzner 2015). It uses NORINE (Caboche et al. 2008), Dictionary of Natural Products, and MarinLit as sources for input chemical structures and then predicts potential MS/MS spectra of these natural products. Matches were accepted with a p value of smaller than 10−10 based on comparison to a shuffled database ensuring no false discoveries. This in silico technique enables the dereplication of metabolites for which high quality reference spectra are not yet available. Despite these tools, the majority of the chemistry in the molecular network still remains unannotated, highlighting an enormous discovery potential for new molecules.
One of the goals of this project was to assess the efficacy of a molecular networking based strategy to find new natural products in the absence of additional information such as genome sequences or bioactivity results. This is important since isolation and structure elucidation remain major bottlenecks in the discovery process, with possible costs for solving a new structure often ranging between 50,000 and 250,000 dollars (Bouslimani et al. 2014). We therefore aimed to build a robust method to prioritize the samples most likely to yield novel metabolites from rapid metabolomic surveys alone. This mode of molecular discovery allows for the efficient analysis of a broad chemical space formed by the more than 900 detected molecular families. As with many large culture collections, metadata and genetic information are sparsely populated. It is therefore desirable to prioritize the chemical space in some self-referential manner. The hypothesis pursued was that if a molecule were found infrequently, it is less likely to have been previously reported. Therefore, the chemistries were visualized through color-coding by the number of strains in the molecular network in which that spectrum had been observed. Those metabolites observed in extracts of only a single strain were highlighted in orange (Fig. 1b, e). About a quarter of all observed chemical entities are only found in a single strain. This supports prior observations of biosynthetic gene cluster diversity among marine actinomycetes (Ziemert et al. 2014). There was however one molecular family in the molecular network that contained over two dozen nodes (3) belonging to a single strain. The molecular family itself contains regular patterns of 186 and 202 Da functional offsets based on the parent masses.
The members of this sizable molecular family (Fig. 2a) were not dereplicated using our computational methods. The most abundant members were recalcitrant to traditional database searches based on parent mass and the inspection of the MS/MS patterns failed to reveal molecular classification features such as evidence of lipids, amino acid or sugar losses. Collectively, these results suggested that this was a unique molecular family in our data set, not represented in our reference databases, and hence possibly representative of a novel group of chemistries. Therefore, the microorganism from the producing well was grown and extracted in large scale. Two of the nodes with m/z values of 971.62 and 1359.87 of the unique molecular family were targeted for further structural elucidation. These two members of the family were observed consistently in high abundance and were targeted for further investigation.
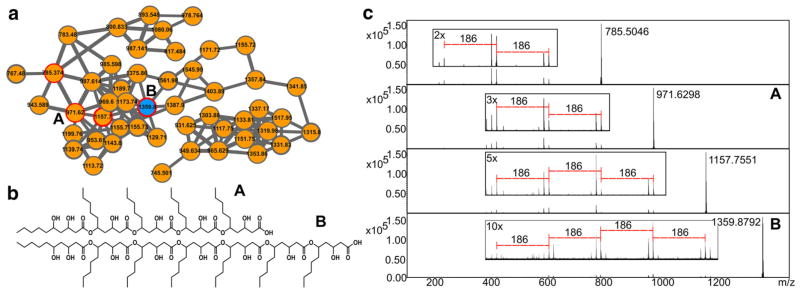
Fig. 2.
The molecular family (a) containing new chemistries maridric acids A and B (b) is composed mainly of nodes derived from a single marine organism (orange). Nodes are labeled with their parent mass and thicker edges between nodes indicate higher spectral similarity score. The four nodes with spectra shown are highlighted with colored borders and the newly identified compounds are labeled in bold. MS/MS spectra (c) of four nodes show the polymeric nature of this molecular family (Color figure online)
The observed ions of m/z 971.62 and 1359.87, corresponding to the maridric acids A and B, respectively, were targeted for the MS-guided isolation. Based on high resolution TOF–MS spectra of the m/z 971.6230 [M+Na]+ ion, the molecular formula was determined as C50H92O16 Na (ppm <2), indicating that the degree of unsaturation number was five. The connectivity of the proton and carbon atoms was established by its 1H–13C HSQC spectrum. (Table S2). Furthermore, 1H–1H COSY and 1H–13C HMBC analyses enabled us to assign the similar five partial structures constituted of 3,5-dihydroxydecanoate as shown in figure S2. The HMBC correlations from H-5, 5′ and 5″ (δ5.01) and H-2′, H-2″ and H-2‴ (δ 2.30 and 2.40) to C-1′, C-1″ and C-1‴ (δ 172.5), and from H-5‴ (δ 5.01) and H-2‴′ (δ 2.35) to C-1‴′ (δ 172.7) showed the connection of the partial structures (Figure S4). Furthermore, the values of carbon chemical shift (δ 66.6, 69.1 and 71.7) suggested that the molecule has six hydroxyl groups at the position 3, 3′, 3″, 3‴, 3‴′ and 5‴′. Similarly, MSn experiments con-firmed a pattern of 186 Da repeating fragment losses consistent with the structure predicted from NMR and MS/MS data. To determine the location of all hydroxyl groups, the molecule was subjected to acetylation followed by LCMS analysis with an ion-trap based instrument. Acetylation of maridric acid A with the acetic anhydride specifically modified alcohol functional groups to yield a six times acetylated derivative with a predicted nominal molecular ion peak at m/z 1224 [M+Na]+ a nominal mass change of 42 m/z. These changes were indeed indicating the introduction of acetyl groups into each hydroxy fatty acid moiety (Figure S3). These MS results strongly supported that maridric acid A has hydroxyl groups at the position 3, 3′, 3″, 3‴, 3‴′ and 5‴′. Finally, the structure as determined by NMR and MS analyses satisfied the degree of unsaturation number and the molecular formula for the pentameric acid. Similarly, NMR and MS analysis (Fig. 2c, Table S2) of the larger molecule showed it to be the heptameric analog as shown in Fig. 2b. These previously undescribed pentameric and heptameric polymers of dihiydroxylated decanoic acid have been designated maridric acids A and B, respectively (Fig. 2b).
Similar structural motifs have previously been reported in the literature from endophytic, and marine fungal organisms, including a trimeric dihydroxydecanoic acid with antibiotic activity against Gram-positive bacteria (Gaskins and Cheung 1986; Cheikh-Ali et al. 2015; Price et al. 2013; Bischoff et al. 2015; Kim et al. 2015). Our MS based chemical survey also revealed other members of that molecular family containing four and six repeats, which represent additional polymeric products (Fig. 2c). The remaining members of the maridric acid family are likely polymeric natural products of the same nature but did not appear to contain these previously reported analogs. Taken together with the large diversity of natural products dereplicated in this study, this discovery highlights the power of MS-based metabolomic techniques to make use the often surprising output of high throughput growths from large culture collections. We show that one can indeed use molecular networking to not only dereplicate known molecules, detect new analogs, quantify the frequency of these molecules in a given strain library, but also to effectively prioritize and discover unexplored chemistries, the cornerstone of many biotechnological advancements.
4 Conclusion
We have shown here that the high throughput growth and extraction of a large culture collection, coupled with untargeted LC–MS/MS metabolomic profiling allows for a large-scale survey of specialized chemistries. Additionally, we find that while global principal component analyses can be great tools for the description of chemical space, molecular networking allows the exploration of this space at the molecular level. By organizing chemical information and linking it to growing spectral reference libraries, analysis with GNPS enabled the rapid dereplication of several molecular entities and provided relatively secure indications of both known and unknown analogs. Finally, we showed the success of a novelty guided approach to strain prioritization, yielding the identification of two previously undescribed molecules. The strength of this technique lies in its untargeted nature. Without reliance on genetic, geographic or other data, large numbers of randomly sampled organisms can be prioritized based on their novel contributions to MS-visible chemical space and guide investigators towards novel molecules. This approach, who’s effectiveness is predicted to grow with scale, provides an effective tool for the untargeted prioritization of microorganisms in varied growth or extraction conditions for the better utilization of large culture collections.
Supplementary Material
Acknowledgments
The authors would like to thank Julia Busch, Natalie Millán, Nastassia Patin, Nick Tuttle for assistance with culturing techniques, William Fenical for strains, as well as Rob Knight and Vanessa Phelan for providing feedback on the manuscript. This work was supported by NIH Grant R01GM085770 (to PJ). DF was supported by NIH Grant T32EB009380. We further acknowledge P41 Grant 5P41GM103484, as well as Bruker and NIH Grant GMS10RR029121 for the support of the shared instrumentation infrastructure that enabled this work.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11306-016-1087-5) contains supplementary material, which is available to authorized users.
Author contributions Writing the manuscript: DJF, PCD. High throughput culturing and data collection: DJF, NK. Isolation and structure elucidation: NK
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflicts of interest.
Human and Animal Research Information This article does not contain any studies with human participants or animals.
References
- Allard P-M, et al. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Analytical Chemistry. 2016 doi: 10.1021/acs.analchem.5b04804. [DOI] [PubMed] [Google Scholar]
- Anonymous. Triphaseco pipeline. n.d Available at: http://triphaseco.com/pipeline/
- Baltz RH, Miao V, Wrigley SK. Natural products to drugs: daptomycin and related lipopeptide antibiotics. Natural Product Reports. 2005;22(6):717–741. doi: 10.1039/b416648p. [DOI] [PubMed] [Google Scholar]
- Bérdy J, View AP. Bioactive microbial metabolites. The Journal of Antibiotics. 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Bischoff KM, et al. Liamocin oil from Aureobasidium pullulans has antibacterial activity with specificity for species of Streptococcus. The Journal of Antibiotics. 2015 doi: 10.1038/ja.2015.39. [DOI] [PubMed] [Google Scholar]
- Blunt JW, et al. Marine natural products. Natural Product Reports. 2014;31(2):160–258. doi: 10.1039/c3np70117d. [DOI] [PubMed] [Google Scholar]
- Bouslimani A, et al. Mass spectrometry of natural products: current, emerging and future technologies. Natural Product Reports. 2014;31(6):718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg RW, et al. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrobial Agents and Chemotherapy. 1979;15(3):361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboche S, et al. NORINE: a database of nonribosomal peptides. Nucleic Acids Research. 2008;36(Database issue):D326–D331. doi: 10.1093/nar/gkm792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheikh-Ali Z, et al. Diversity of exophillic acid derivatives in strains of an endophytic Exophiala sp. Phytochemistry. 2015;118:83–93. doi: 10.1016/j.phytochem.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Chen C, et al. Halymecins, new antimicroalgal substances produced by fungi isolated from marine algae. The Journal of Antibiotics. 1996;49(10):998–1005. doi: 10.7164/antibiotics.49.998. [DOI] [PubMed] [Google Scholar]
- Choi H, et al. Honaucins A-C, potent inhibitors of inflammation and bacterial quorum sensing: synthetic derivatives and structure-activity relationships. CHEMISTRY & BIOLOGY. 2012;19(5):589–598. doi: 10.1016/j.chembiol.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WB, et al. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics. 2012;9(S1):44–66. [Google Scholar]
- Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nature Chemical Biology. 2006;2(12):666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- Fenical W, et al. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorganic & Medicinal Chemistry. 2009;17(6):2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzæ. British Journal of Experimental Pathology. 1929;10(3):226. [Google Scholar]
- Frank AM, et al. Clustering millions of tandem mass spectra. Journal of Proteome Research. 2008;7(1):113–122. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins JE, Cheung PJ. Exophiala pisciphila. A study of its development. Mycopathologia. 1986;93(3):173–184. doi: 10.1007/BF00443521. [DOI] [PubMed] [Google Scholar]
- Gerwick WH, Moore BS. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chemistry & Biology. 2012;19(1):85–98. doi: 10.1016/j.chembiol.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AC, et al. ECMDB: the E. coli metabolome database. Nucleic Acids Research. 2013;41(Database issue):D625–D630. doi: 10.1093/nar/gks992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai H, et al. MassBank: a public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry. 2010;45(7):703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- Hou Y, et al. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Analytical Chemistry. 2012;84(10):4277–4283. doi: 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, et al. Statistical research on the bioactivity of new marine natural products discovered during the 28 years from 1985 to 2012. Marine Drugs. 2015;13(1):202–221. doi: 10.3390/md13010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. The distribution of the flora in the alpine zone.1. New Phytologist. 1912;11(2):37–50. [Google Scholar]
- Kallmeyer J, et al. Global distribution of microbial abundance and biomass in subseafloor sediment. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(40):16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg JJ, et al. Biochemometrics for natural products research: comparison of data analysis approaches and application to identification of bioactive compounds. Journal of Natural Products. 2016 doi: 10.1021/acs.jnatprod.5b01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LW, et al. Local genomic adaptation of coral reef-associated microbiomes to gradients of natural variability and anthropogenic stressors. Proceedings of the National Academy of Sciences. 2014;111(28):10227–10232. doi: 10.1073/pnas.1403319111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten RD, et al. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nature Chemical Biology. 2011;7(11):794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lee IK, Yun BS. A novel biosurfactant produced by Aureobasidium pullulans L3-GPY from a tiger lily wild flower, Lilium lancifolium Thunb. PloS One. 2015;10(4):e0122917. doi: 10.1371/journal.pone.0122917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. LC-MS/MS profiling-based secondary metabolite screening of Myxococcus xanthus. Journal of Microbiology and Biotechnology. 2009;19(1):51–54. [PubMed] [Google Scholar]
- Kinkel LL, et al. Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among Streptomycetes. The ISME Journal. 2014;8(2):249–256. doi: 10.1038/ismej.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitgaard A, et al. Combining stable isotope labeling and molecular networking for biosynthetic pathway characterization. Analytical Chemistry. 2015;87(13):6520–6526. doi: 10.1021/acs.analchem.5b01934. [DOI] [PubMed] [Google Scholar]
- Kurita KL, Glassey E, Linington RG. Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries. Proceedings of the National Academy of Sciences. 2015;112(39):11999–12004. doi: 10.1073/pnas.1507743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, et al. ChemGPS-NP: tuned for navigation in biologically relevant chemical space. Journal of Natural Products. 2007;70(5):789–794. doi: 10.1021/np070002y. [DOI] [PubMed] [Google Scholar]
- Marrone TJ, Merz KMJ. Molecular recognition of K+ and Na+ by valinomycin in methanol. Journal of the American Chemical Society. 1995;117(2):779–791. [Google Scholar]
- Mohimani H, Pevzner PA. Dereplication, sequencing and identification of peptidic natural products: from genome mining to peptidogenomics to spectral networks. Natural Product Reports. 2015;33(1):73–86. doi: 10.1039/c5np00050e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaser R, Luesch H. Marine natural products: a new wave of drugs? Future Medicinal Chemistry. 2011;3(12):1475–1489. doi: 10.4155/fmc.11.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DD, et al. MS/MS networking guided analysis of molecule and gene cluster families. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(28):E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KF, et al. Dereplication of microbial natural products by LC-DAD-TOFMS. Journal of Natural Products. 2011;74(11):2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- Price NPJ, et al. Structural characterization of novel extracellular liamocins (mannitol oils) produced by Aureobasidium pullulans strain NRRL 50380. Carbohydrate Research. 2013;370:24–32. doi: 10.1016/j.carres.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Quinn RA, Alexandrov T. The community ecology of microbial molecules. Journal of Chemical Ecology. 2014;40(11–12):1161–1162. doi: 10.1007/s10886-014-0528-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P, et al. New drugs from marine organisms in Alzheimer’s disease. Marine Drugs. 2015;14(1):5. doi: 10.3390/md14010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: a plant-specific MS/MS-based data resource and database. Phytochemistry. 2012;82:38–45. doi: 10.1016/j.phytochem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Sehgal SN, Baker H, Vézina C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. The Journal of Antibiotics. 1975;28(10):727–732. doi: 10.7164/antibiotics.28.727. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. Culture collections over the world. International Microbiology. 2003;6(2):95–100. doi: 10.1007/s10123-003-0114-3. [DOI] [PubMed] [Google Scholar]
- Subramani R, Aalbersberg W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiological Research. 2012;167(10):571–580. doi: 10.1016/j.micres.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, et al. XCMS Online: A web-based platform to process untargeted metabolomic data. Analytical Chemistry. 2012;84(11):5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y, et al. EMPeror: A tool for visualizing high-throughput microbial community data. GigaScience. 2013;2(1):16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino M, et al. Discovering and deciphering the pathogenic and probiotic activities from the bacterial colibactin pathway. Planta Medica. 2015;81(11):IL40. [Google Scholar]
- Watrous J, et al. Mass spectral molecular networking of living microbial colonies. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(26):E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proceedings of the National Academy of Sciences. 1998;95(12):6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, et al. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Research. 2013;41(Database issue):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nature Biotechnology. doi: 10.1038/nbt.3597. (accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, et al. Molecular networking as a dereplication strategy. Journal of Natural Products. 2013;76(9):1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, et al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(12):E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.