Fig. 1.
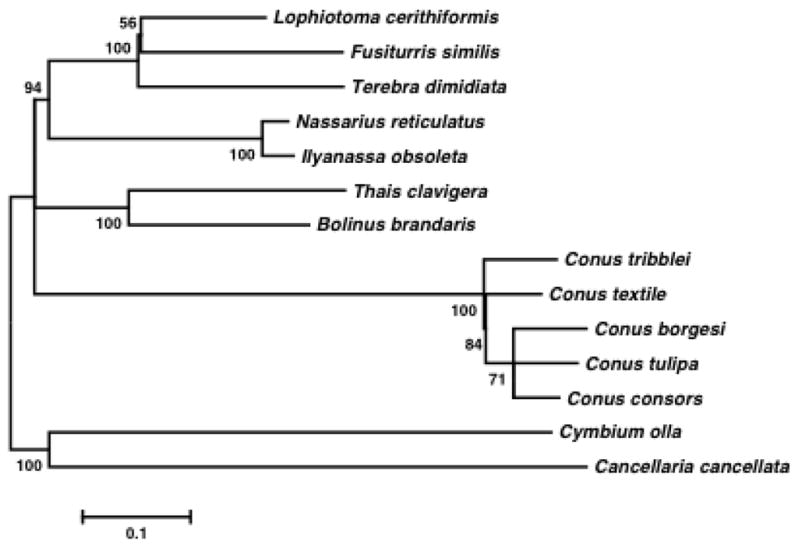
Phylogenetic relationship within Neogastropods. The evolutionary history was inferred using the Maximum likelihood method based on the General Reversible Mitochondrial+Freq model from a single concatenated data set of deduced amino acid sequences of 13 mitochondrial protein-coding genes. A discrete Gamma distribution was used to model evolutionary rate differences among cites. The rate variation model allowed for some sites to be evolutionary invariable. The bootstrap values are indicated on the nodes. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA5 (Tamura et al. 2011). Conus tulipa (KR006970), Conus borgesi (EU827198), Conus consors (KF887950), Conus textile (DQ862058), Terebra dimidiata (EU827196), Fusiturris similis (EU827197), Lophiotoma cerithiformis (DQ284754), Cancellaria cancellata (EU827195), Cymbium olla (EU827199), Bolinus brandaris (EU827194), Thais clavigera (DQ159954), Ilyanassa obsoleta (DQ238598) and Nassarius reticulatus (EU827201).
