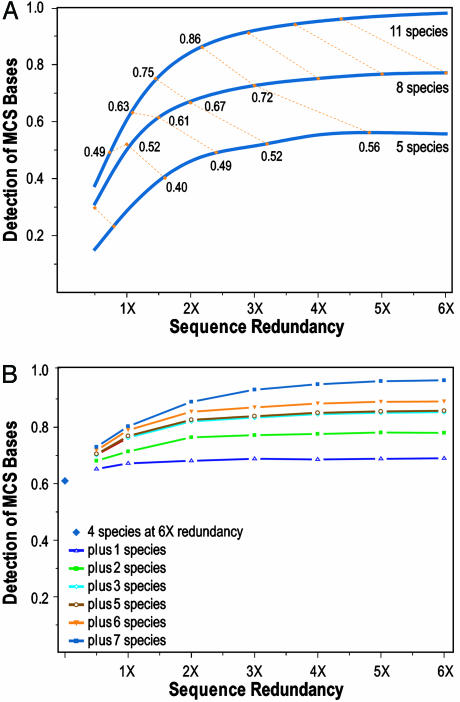
Fig. 4.
Identification of MCSs with various levels of sequence redundancy. (A) The finished sequences of ENCODE region ENm001 (7, 18, 22) from 11 mammalian species were used to identify a reference set of MCSs. MCS detection was then repeated with subsets of the data providing several lower levels of sequence redundancy (using both assembled sequence contigs and unassembled reads) for different subsets of mammals. The threshold was set to ensure 97% specificity for detecting the reference set of MCS bases. The performance of detecting MCSs with 5 (lemur, dog, horse, hedgehog, and mouse), 8 (the previous 5 plus pig, armadillo, and rabbit), or 11 (the previous 8 plus cat, cow, and rat) species is depicted by three smoothed curves statistically fit to the actual data (see supporting information). The yellow dotted lines connect “iso-read” equivalents (see text for details) for 5, 8, and 11 mammals, calculated for discrete increments of redundancy for 8 mammals (0.5- through 6-fold redundancy), with the indicated numbers reflecting the sensitivity of MCS detection for that data point. (B) Analogous studies were performed with region ENm005, but starting with 6-fold redundant sequence from 4 species for which whole-genome sequence is already available (dog, rat, mouse, and chicken) and then adding sequence from 7 additional species [in the order cat, cow, pig, fugu plus Tetraodon (pufferfish), galago, and baboon].