Fig. 1. CpG distribution in the SNCA 5′ regulatory region.
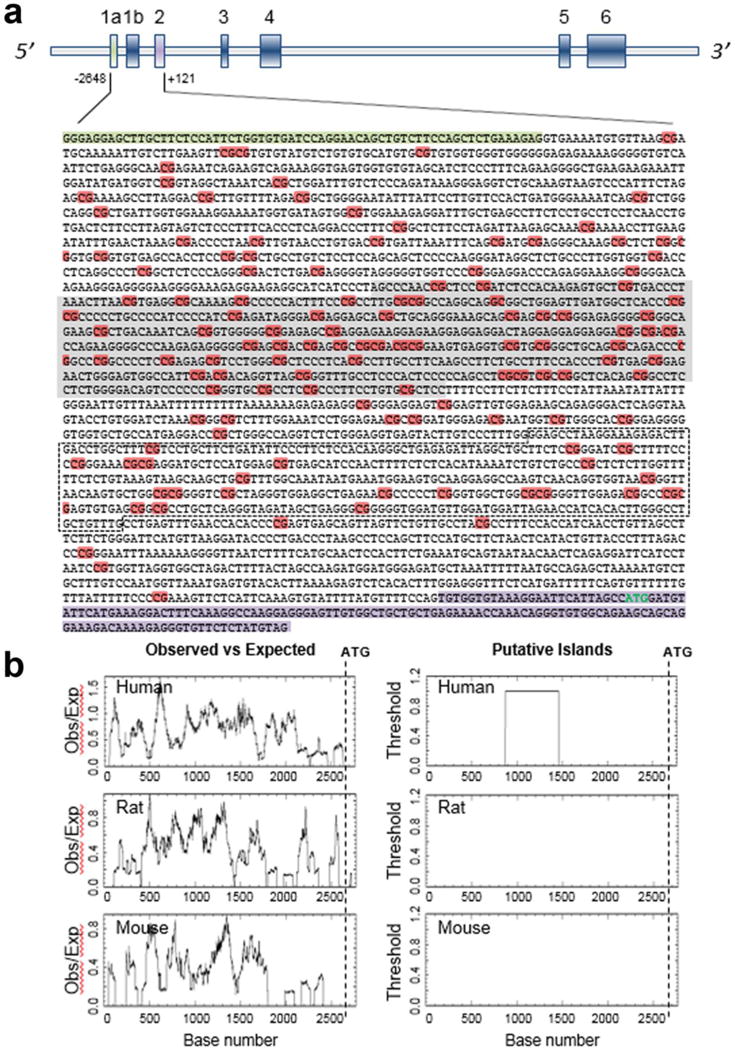
Human SNCA gene has 6 exons with two alternative non-coding exon 1s. The translation start codon is located in exon 2. The vertical bars represent each exon. The sequence from +121 to −2,648 with respect to ATG (+1) (marked in green), encompassing both non-coding exons, intron1 and exon 2, shows very high density of CG bases as highlighted in orange. The presence of the CpG island was predicted using the CpGPLOT program (http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/) with default parameter; the predicted island of 591 base pairs is highlighted in grey. The black dashed marked box represents the region in intron1 whose CpG methylation status has been analyzed by most of the studies (A). Comparative CpG density between human, mouse and rat α-SYN gene (−2,648 to +1) was done using the same program as mentioned above. Analysis revealed a significant difference in CpG base distribution between human and other species when the same region of the gene was tested. Only human showed the presence of a CpG island >200 bp, and the ratio between the observed CpG distribution over expected region ranged mostly in between 1.0 to 1.5. The ratio went down to 0.8 when the other two species were analyzed for CpG distribution for the same region (B). The cut-off for the ratio was set at 0.6.
