Fig. 2. Differential epigenetic landscape of SNCA promoter in different cell lines.
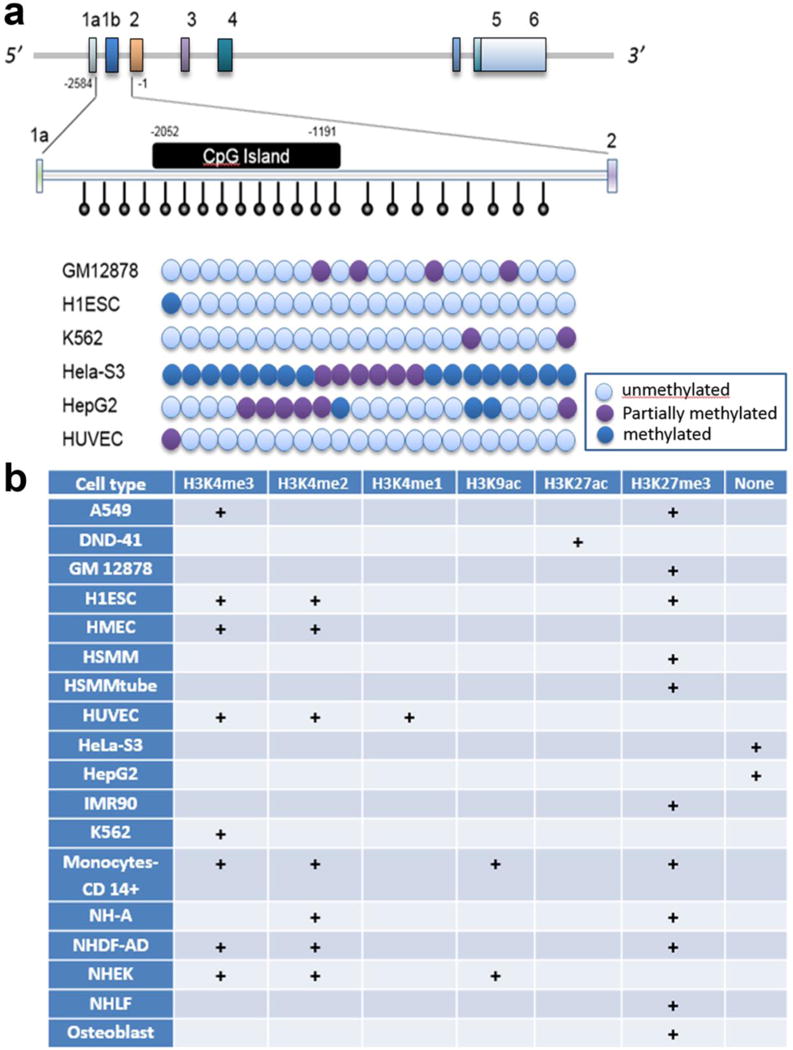
SNCA promoter and upstream region was analyzed for methylation status of its CpG rich area and occupancy of different transcription related histone marks. The entire analyses is based on the data from ENCODE using UCSC genome browser. ENCODE data showed the presence of a CpG island of around 861 bp with the starting base slightly upstream when compared to that of our analysis using CpGPLOT program as shown in Figure 1 (−2052 to −1191 bp in case of ENCODE analysis Vs −1777 to −1187 bp using CpGPLOT program; all positions are designated considering ATG as +1) (A). Twenty-two CpG sites distributed over the SNCA promoter/intron1 area were analyzed using 450K methylation bead array in the ENCODE database. The relative position of those 22 CpG sites within exon 1a to exon 2 is shown by downward vertical bars with round head. The methylation statuses of all 22 CpGs for 6 different cell lines are summarized in the figure: The blue ball represents unmethylated cytosines, pink represents methylated cytosines, and purple shows partially methylated cytosines. The ENCODE database showed that GM12878, H1ESC, HUVEC and K562 remains majorly unmethylated for respective CpGs, HeLa-S3 exhibits a heavily methylated pattern, and HepG2 cells demonstrates partial methylation across the 22 CpG sites. Different histone PTMs associated with transcriptional activity were also assessed from ENCODE database for similar areas of SNCA across different cell lines, as demonstrated by the matrix representation (B). H3K4me1/2/3, H3K9ac, H3K27ac represent active state of chromatin where as H3K27me3 and absence of any histone PTMs (designated as “None”) are associated with inactive state of chromatin. Each cell type shows possession of one or more different types of histone PTMs in similar or slightly different areas in the promoter, and their presence is designated by +. A549, human adenocarcinomic alveolar basal epithelial cell line; DND-41, human T cell leukemic cell line; GM12878, human lymphoblastoid cell line; H1ESC, human embryonic cell line; HMEC, human mammary epithelial cell line; HSMC, human skeletal muscle and myoblast; HSMMtube, skeletal muscle myotubes differentiated from HSMM cell line; HUVEC, human umbilical vein endothelial cells; HeLa-S3, clonal derivative from HeLa cells; HepG2, human liver hepatocellular cells; IMR90, human fetal lung cell line; K562, chronic myelogenous leukemia cell line; NH-A, normal human astrocyte cells; NHDF-AD, normal human dermal fibroblast cells; NHEK, normal human epidermal keratinocytes; NHLF, normal human fibroblast cells.
